The Stronger Downregulation of in vitro and in vivo Innate Antiviral Responses by a Very Virulent Strain of Infectious Bursal Disease Virus (IBDV), Compared to a Classical Strain, Is Mediated, in Part, by the VP4 Protein
- PMID: 32582573
- PMCID: PMC7296162
- DOI: 10.3389/fcimb.2020.00315
The Stronger Downregulation of in vitro and in vivo Innate Antiviral Responses by a Very Virulent Strain of Infectious Bursal Disease Virus (IBDV), Compared to a Classical Strain, Is Mediated, in Part, by the VP4 Protein
Abstract
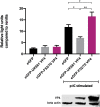
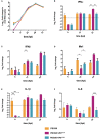
IBDV is economically important to the poultry industry. Very virulent (vv) strains cause higher mortality rates than other strains for reasons that remain poorly understood. In order to provide more information on IBDV disease outcome, groups of chickens (n = 18) were inoculated with the vv strain, UK661, or the classical strain, F52/70. Birds infected with UK661 had a lower survival rate (50%) compared to F52/70 (80%). There was no difference in peak viral replication in the bursa of Fabricius (BF), but the expression of chicken IFNα, IFNβ, MX1, and IL-8 was significantly lower in the BF of birds infected with UK661 compared to F52/70 (p < 0.05) as quantified by RTqPCR, and this trend was also observed in DT40 cells infected with UK661 or F52/70 (p < 0.05). The induction of expression of type I IFN in DF-1 cells stimulated with polyI:C (measured by an IFN-β luciferase reporter assay) was significantly reduced in cells expressing ectopic VP4 from UK661 (p < 0.05), but was higher in cells expressing ectopic VP4 from F52/70. Cells infected with a chimeric recombinant IBDV carrying the UK661-VP4 gene in the background of PBG98, an attenuated vaccine strain that induces high levels of innate responses (PBG98-VP4UK661) also showed a reduced level of IFNα and IL-8 compared to cells infected with a chimeric virus carrying the F52/70-VP4 gene (PBG98-VP4F52/70) (p < 0.01), and birds infected with PBG98-VP4UK661 also had a reduced expression of IFNα in the BF compared to birds infected with PBG98-VP4F52/70 (p < 0.05). Taken together, these data demonstrate that UK661 induced the expression of lower levels of anti-viral type I IFN and proinflammatory genes than the classical strain in vitro and in vivo and this was, in part, due to strain-dependent differences in the VP4 protein.
Keywords: IBDV; VP4; cytokines; inflammation; type I IFN; virulence.
Copyright © 2020 Dulwich, Asfor, Gray, Giotis, Skinner and Broadbent.
Figures




References
-
- Bayliss C. D., Spies U., Shaw K., Peters R. W., Papageorgiou A., Muller H., et al. (1990). A comparison of the sequences of segment A of four infectious bursal disease virus strains and identification of a variable region in VP2. J. Gen. Virol. 71 (Pt 6), 1303–1312. 10.1099/0022-1317-71-6-1303 - DOI - PubMed
-
- Brown M. D., Green P., Skinner M. A. (1994). VP2 sequences of recent European ‘very virulent’ isolates of infectious bursal disease virus are closely related to each other but are distinct from those of ‘classical’ strains. J. Gen. Virol. 75 (Pt 3), 675–680. 10.1099/0022-1317-75-3-675 - DOI - PubMed
-
- Campbell E. A., Reddy V. R. A. P., Gray A. G., Wells J., Simpson J., Skinner M. A., et al. (2020). Discrete virus factories form in the cytoplasm of cells co-infected with two replication competent tagged reporter birnaviruses, that subsequently coalesce over time. J. Virol. 10.1128/JVI.02107-19. [Epub ahead of print]. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- 104771/Z/14/Z/WT_/Wellcome Trust/United Kingdom
- BBS/E/I/00002115/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BBS/E/I/00007038/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/K002465/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BBS/E/I/00007039/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Miscellaneous

