Rumen Microbiota Distribution Analyzed by High-Throughput Sequencing After Oral Doxycycline Administration in Beef Cattle
- PMID: 32582771
- PMCID: PMC7280444
- DOI: 10.3389/fvets.2020.00251
Rumen Microbiota Distribution Analyzed by High-Throughput Sequencing After Oral Doxycycline Administration in Beef Cattle
Abstract
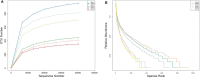
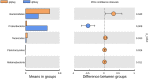
The beef cattle rumen is a heterogenous microbial ecosystem that is necessary for the host to digest food and support growth. The importance of the rumen microbiota (RM) is also widely recognized for its critical roles in metabolism and immunity. The level of health is indicated by a dynamic RM distribution. We performed high-throughput sequencing of the bacterial 16S rRNA gene to compare microbial populations between rumens in beef cattle with or without doxycycline treatment to assess dynamic microbiotic shifts following antibiotic administration. The results of the operational taxonomic unit analysis and alpha and beta diversity calculations showed that doxycycline-treated beef cattle had lower species richness and bacterial diversity than those without doxycycline. Bacteroidetes was the predominant phylum in rumen samples without doxycycline, while Proteobacteria was the governing phylum in the presence of doxycycline. On the family level, the top three predominant populations in group qlqlwy (not treated with doxycycline) were Prevotellaceae, Lachnospiraceae, and Ruminococcaceae, compared to Xanthomonadaceae, Prevotellaceae, and Rikenellaceae in group qlhlwy (treated with doxycycline). At the genus level, the top predominant population in group qlqlwy was unidentified_Prevotellaceae. However, in group qlhlwy, the top predominant population was Stenotrophomonas. The results revealed significant RM differences in beef cattle with or without doxycycline. Oral doxycycline may induce RM composition differences, and bacterial richness may also influence corresponding changes that could guide antibiotic use in adult ruminants. This study is the first to assess microbiota distribution in beef cattle rumen after doxycycline administration.
Keywords: MiSeq sequencing; bacterial richness; beef cattle; doxycycline; dysbacteriosis; oral antibiotics; rumen microbiota.
Copyright © 2020 Chen, Cheng, Xu, Wang, Xia and Hu.
Figures


References
-
- Gruninger RJ, Puniya AK, Callaghan TM, Edwards JE, Youssef N, Dagar SS, et al. Anaerobic fungi (phylum Neocallimastigomycota): advances in understanding their taxonomy, life cycle, ecology, role and biotechnological potential. FEMS Microbiol Ecol. (2014) 90:1–17. 10.1111/1574-6941.12383 - DOI - PubMed
LinkOut - more resources
Full Text Sources

