A Role for Taok2 in Listeria monocytogenes Vacuolar Escape
- PMID: 32582947
- PMCID: PMC8922001
- DOI: 10.1093/infdis/jiaa367
A Role for Taok2 in Listeria monocytogenes Vacuolar Escape
Abstract
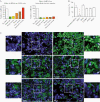
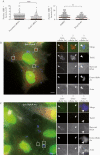
The bacterial pathogen Listeria monocytogenes invades host cells, ruptures the internalization vacuole, and reaches the cytosol for replication. A high-content small interfering RNA (siRNA) microscopy screen allowed us to identify epithelial cell factors involved in L. monocytogenes vacuolar rupture, including the serine/threonine kinase Taok2. Kinase activity inhibition using a specific drug validated a role for Taok2 in favoring L. monocytogenes cytoplasmic access. Furthermore, we showed that Taok2 recruitment to L. monocytogenes vacuoles requires the presence of pore-forming toxin listeriolysin O. Overall, our study identified the first set of host factors modulating L. monocytogenes vacuolar rupture and cytoplasmic access in epithelial cells.
Keywords: Listeria monocytogenes; STE1-like kinase; Taok2; siRNA screen; vacuolar escape.
© The Author(s) 2020. Published by Oxford University Press for the Infectious Diseases Society of America.
Figures
References
-
- Pizarro-Cerdá J, Cossart P. Microbe profile: Listeria monocytogenes: a paradigm among intracellular bacterial pathogens. Microbiology 2019; 165:719–21. - PubMed
-
- Mellouk N, Weiner A, Aulner N, et al. Shigella subverts the host recycling compartment to rupture its vacuole. Cell Host Microbe 2014; 16:517–30. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

