MicroRNA‑103 modulates tumor progression by targeting KLF7 in non‑small cell lung cancer
- PMID: 32582959
- PMCID: PMC7387085
- DOI: 10.3892/ijmm.2020.4649
MicroRNA‑103 modulates tumor progression by targeting KLF7 in non‑small cell lung cancer
Retraction in
-
[Retracted] MicroRNA‑103 modulates tumor progression by targeting KLF7 in non‑small cell lung cancer.Int J Mol Med. 2026 Jan;57(1):24. doi: 10.3892/ijmm.2025.5695. Epub 2025 Nov 21. Int J Mol Med. 2026. PMID: 41268601 Free PMC article.
Abstract
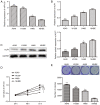
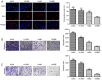
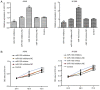
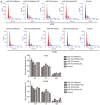
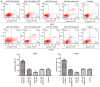
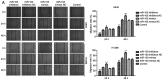
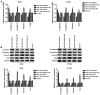
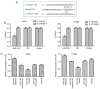
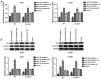
Numerous studies have identified that microRNAs (miRs) play a crucial role in the tumorigenesis of non‑small cell lung cancer (NSCLC). However, to the best of our knowledge, the physiological function of miR‑103 in NSCLC is not fully understood. Experiments in the present study revealed that miR‑103 expression was increased in NSCLC cell lines. In addition, a series of methods, including MTT, colony formation, 5‑ethynyl‑2'‑deoxyuridine, Transwell, wound healing, flow cytometric, reverse transcription‑quantitative PCR and western blot assays, were performed, which revealed that overexpression of miR‑103 enhanced cell growth, migration, invasion and epithelial‑mesenchymal transition (EMT), and suppressed apoptosis of A549 and H1299 cells. Additionally, a dual‑luciferase reporter assay indicated that miR‑103 directly targets the 3'‑untranslated region of Kruppel‑like factor 7 (KLF7), and KLF7 expression was negatively regulated by miR‑103 expression. Furthermore, the present findings demonstrated that miR‑103 promoted EMT via regulating the Wnt/β‑catenin signaling pathway in NSCLC. Collectively, the current results demonstrated that miR‑103 serves a tumorigenesis role in NSCLC development by targeting KLF7, at least partly via the Wnt/β‑catenin signaling pathway. Consequently, these findings indicated that miR‑103/KLF7/Wnt/β‑catenin may provide a novel insight into underlying biomarkers for improving the diagnosis and treatment of NSCLC.
Figures








References
-
- Goldstraw P, Chansky K, Crowley J, Porta RR, Asamura H, Eberhardt WE, Nicholson AG, Groome P, Mitchell A, Bolejack V, et al. The IASLC lung cancer staging project: Proposals for revision of the tnm stage groupings in the forthcoming (Eighth) edition of the TNM classification for lung cancer. J Thorac Oncol. 2016;11:39–51. doi: 10.1016/j.jtho.2015.09.009. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

