Early M-Protein Dynamics Predicts Progression-Free Survival in Patients With Relapsed/Refractory Multiple Myeloma
- PMID: 32583948
- PMCID: PMC7719372
- DOI: 10.1111/cts.12836
Early M-Protein Dynamics Predicts Progression-Free Survival in Patients With Relapsed/Refractory Multiple Myeloma
Abstract
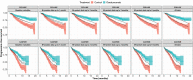
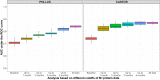
This study aimed to predict long-term progression-free survival (PFS) using early M-protein dynamic measurements in patients with relapsed/refractory multiple myeloma (MM). The PFS was modeled based on dynamic M-protein data from two phase III studies, POLLUX and CASTOR, which included 569 and 498 patients with relapsed/refractory MM, respectively. Both studies compared active controls (lenalidomide and dexamethasone, and bortezomib and dexamethasone, respectively) alone vs. in combination with daratumumab. Three M-protein dynamic features from the longitudinal M-protein data were evaluated up to different time cutoffs (1, 2, 3, and 6 months). The abilities of early M-protein dynamic measurements to predict the PFS were evaluated using Cox proportional hazards survival models. Both univariate and multivariable analyses suggest that maximum reduction of M-protein (i.e., depth of response) was the most predictive of PFS. Despite the statistical significance, the baseline covariates provided very limited predictive value regarding the treatment effect of daratumumab. However, M-protein dynamic features obtained within the first 2 months reasonably predicted PFS and the associated treatment effect of daratumumab. Specifically, the areas under the time-varying receiver operating characteristic curves for the model with the first 2 months of M-protein dynamic data were ~ 0.8 and 0.85 for POLLUX and CASTOR, respectively. Early M-protein data within the first 2 months can provide a prospective and reasonable prediction of future long-term clinical benefit for patients with MM.
© 2020 The Authors. Clinical and Translational Science published by Wiley Periodicals LLC on behalf of the American Society for Clinical Pharmacology and Therapeutics.
Conflict of interest statement
X.S.X., T.P., J.U., K.B., Q.M., S.S., and H.Z. are employees of Janssen Research & Development, LLC. X.S.X., T.P., J.U., K.B., Q.M., S.S., and H.Z. own stock in Johnson & Johnson. K.W. reports research funding from Janssen, Amgen, Sanofi, and Celgene Corporation; honoraria from Amgen, Bristol‐Myers Squibb, Celgene Corporation, Janssen, Novartis, Onyx, and Takeda; and advisory board membership for Adaptive Biotech, Amgen, Bristol‐Myers Squibb, Celgene Corporation, Janssen, Novartis, Onyx, and Takeda. M.V.M. received honoraria from and consulted for Janssen, Celgene, Takeda, and Amgen. A.S. received research funding and personal fees from Janssen Cilag. P.S. reports research funding from Janssen, Celgene, Amgen, and Takeda; received honoraria from Janssen, Celgene, Amgen, and Takeda; and provided expert testimony for Pharma Mar. M.A.D. consulted for Amgen, Janssen‐Cilag, and Takeda; received travel expenses from Janssen‐Ortho and Genesis Pharmaceuticals; received honoraria from Amgen, Novartis, Celgene, Takeda, Genesis Pharmaceuticals, Janssen‐Cilag, and Bristol‐Myers Squibb; and received research funding from Janssen‐Cilag and Amgen. S.Z.U. consulted for Abbvie, GlaxoSmithKline, Celgene, Amgen/Onyx, Takeda/Millennium, Sanofi, Seattle Genetics, Skyline, Merck, and Janssen; received research funding from Celgene, Amgen/Onyx, Takeda/Millennium, Sanofi, Seattle Genetics, Skyline, Merck, Janssen, Array BioPharma, and Pharmacyclics; served on speakers bureaus for Celgene, Amgen, Janssen, Sanofi, and Takeda; and received travel expenses from Janssen, Celgene, Amgen, and Takeda. N.J.B. received honoraria and travel expenses from Celgene, Takeda, Janssen, and Amgen; served on advisory boards for Celgene, Takeda, Janssen, and Amgen; served on speaker’s bureaus for Celgene, Janssen, and Amgen; and received research funding from and provided expert testimony to Celgene and Janssen. All other authors declared no competing interests for this work.
Figures



References
-
- de Weers, M. et al Daratumumab, a novel therapeutic human CD38 monoclonal antibody, induces killing of multiple myeloma and other hematological tumors. J. Immunol. 186, 1840–1848 (2011). - PubMed
-
- Overdijk, M. B. et al The therapeutic CD38 monoclonal antibody daratumumab induces programmed cell death via Fcgamma receptor‐mediated cross‐linking. J. Immunol. 197, 807–813 (2016). - PubMed
-
- Lammerts van Bueren, J. et al Direct in vitro comparison of daratumumab with surrogate analogs of CD38 antibodies MOR03087, SAR650984 and Ab79. Blood 124, 3474 (2014).
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

