Cbl and Cbl-b control the germinal center reaction by facilitating naive B cell antigen processing
- PMID: 32584413
- PMCID: PMC7478728
- DOI: 10.1084/jem.20191537
Cbl and Cbl-b control the germinal center reaction by facilitating naive B cell antigen processing
Abstract
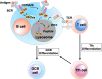
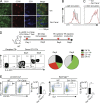
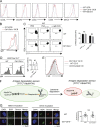
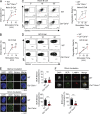
Antigen uptake and presentation by naive and germinal center (GC) B cells are different, with the former expressing even low-affinity BCRs efficiently capture and present sufficient antigen to T cells, whereas the latter do so more efficiently after acquiring high-affinity BCRs. We show here that antigen uptake and processing by naive but not GC B cells depend on Cbl and Cbl-b (Cbls), which consequently control naive B and cognate T follicular helper (Tfh) cell interaction and initiation of the GC reaction. Cbls mediate CD79A and CD79B ubiquitination, which is required for BCR-mediated antigen endocytosis and postendocytic sorting to lysosomes, respectively. Blockade of CD79A or CD79B ubiquitination or Cbls ligase activity is sufficient to impede BCR-mediated antigen processing and GC development. Thus, Cbls act at the entry checkpoint of the GC reaction by promoting naive B cell antigen presentation. This regulation may facilitate recruitment of naive B cells with a low-affinity BCR into GCs to initiate the process of affinity maturation.
© 2020 Li et al.
Conflict of interest statement
Disclosures: The authors declare no competing interests exist.
Figures




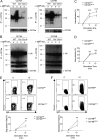
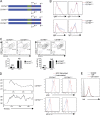


Comment in
-
Cbls boost B cells.J Exp Med. 2020 Sep 7;217(9):e20201105. doi: 10.1084/jem.20201105. J Exp Med. 2020. PMID: 32813871 Free PMC article.
Similar articles
-
Cbl Ubiquitin Ligases Control B Cell Exit from the Germinal-Center Reaction.Immunity. 2018 Mar 20;48(3):530-541.e6. doi: 10.1016/j.immuni.2018.03.006. Immunity. 2018. PMID: 29562201
-
The Syk-binding ubiquitin ligase c-Cbl mediates signaling-dependent B cell receptor ubiquitination and B cell receptor-mediated antigen processing and presentation.J Biol Chem. 2012 May 11;287(20):16636-44. doi: 10.1074/jbc.M112.357640. Epub 2012 Mar 27. J Biol Chem. 2012. PMID: 22451666 Free PMC article.
-
Deficiency of CBL and CBLB ubiquitin ligases leads to hyper T follicular helper cell responses and lupus by reducing BCL6 degradation.Immunity. 2024 Jul 9;57(7):1603-1617.e7. doi: 10.1016/j.immuni.2024.04.023. Epub 2024 May 17. Immunity. 2024. PMID: 38761804
-
BCR Affinity Influences T-B Interactions and B Cell Development in Secondary Lymphoid Organs.Front Immunol. 2021 Jul 26;12:703918. doi: 10.3389/fimmu.2021.703918. eCollection 2021. Front Immunol. 2021. PMID: 34381455 Free PMC article. Review.
-
Regulation of immune system development and function by Cbl-mediated ubiquitination.Immunol Rev. 2019 Sep;291(1):123-133. doi: 10.1111/imr.12789. Immunol Rev. 2019. PMID: 31402498 Review.
Cited by
-
Immune dysregulation caused by homozygous mutations in CBLB.J Clin Invest. 2022 Oct 17;132(20):e154487. doi: 10.1172/JCI154487. J Clin Invest. 2022. PMID: 36006710 Free PMC article.
-
Csk restrains BCR-mediated ROS production and contributes to germinal center selection and affinity maturation.J Exp Med. 2024 Jul 1;221(7):e20231996. doi: 10.1084/jem.20231996. Epub 2024 May 16. J Exp Med. 2024. PMID: 38753246 Free PMC article.
-
Role of B cells as antigen presenting cells.Front Immunol. 2022 Sep 8;13:954936. doi: 10.3389/fimmu.2022.954936. eCollection 2022. Front Immunol. 2022. PMID: 36159874 Free PMC article. Review.
-
The Ins and Outs of Antigen Uptake in B cells.Front Immunol. 2022 Apr 26;13:892169. doi: 10.3389/fimmu.2022.892169. eCollection 2022. Front Immunol. 2022. PMID: 35572544 Free PMC article. Review.
-
B-cell intrinsic regulation of antibody mediated immunity by histone H2A deubiquitinase BAP1.Front Immunol. 2024 Mar 11;15:1353138. doi: 10.3389/fimmu.2024.1353138. eCollection 2024. Front Immunol. 2024. PMID: 38529289 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

