Antibody tests for identification of current and past infection with SARS-CoV-2
- PMID: 32584464
- PMCID: PMC7387103
- DOI: 10.1002/14651858.CD013652
Antibody tests for identification of current and past infection with SARS-CoV-2
Update in
-
Antibody tests for identification of current and past infection with SARS-CoV-2.Cochrane Database Syst Rev. 2022 Nov 17;11(11):CD013652. doi: 10.1002/14651858.CD013652.pub2. Cochrane Database Syst Rev. 2022. PMID: 36394900 Free PMC article.
Abstract
Background: The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) virus and resulting COVID-19 pandemic present important diagnostic challenges. Several diagnostic strategies are available to identify current infection, rule out infection, identify people in need of care escalation, or to test for past infection and immune response. Serology tests to detect the presence of antibodies to SARS-CoV-2 aim to identify previous SARS-CoV-2 infection, and may help to confirm the presence of current infection.
Objectives: To assess the diagnostic accuracy of antibody tests to determine if a person presenting in the community or in primary or secondary care has SARS-CoV-2 infection, or has previously had SARS-CoV-2 infection, and the accuracy of antibody tests for use in seroprevalence surveys.
Search methods: We undertook electronic searches in the Cochrane COVID-19 Study Register and the COVID-19 Living Evidence Database from the University of Bern, which is updated daily with published articles from PubMed and Embase and with preprints from medRxiv and bioRxiv. In addition, we checked repositories of COVID-19 publications. We did not apply any language restrictions. We conducted searches for this review iteration up to 27 April 2020.
Selection criteria: We included test accuracy studies of any design that evaluated antibody tests (including enzyme-linked immunosorbent assays, chemiluminescence immunoassays, and lateral flow assays) in people suspected of current or previous SARS-CoV-2 infection, or where tests were used to screen for infection. We also included studies of people either known to have, or not to have SARS-CoV-2 infection. We included all reference standards to define the presence or absence of SARS-CoV-2 (including reverse transcription polymerase chain reaction tests (RT-PCR) and clinical diagnostic criteria).
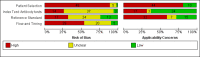
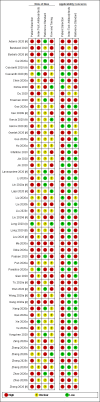
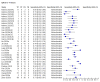
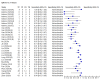
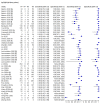
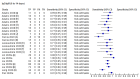
Data collection and analysis: We assessed possible bias and applicability of the studies using the QUADAS-2 tool. We extracted 2x2 contingency table data and present sensitivity and specificity for each antibody (or combination of antibodies) using paired forest plots. We pooled data using random-effects logistic regression where appropriate, stratifying by time since post-symptom onset. We tabulated available data by test manufacturer. We have presented uncertainty in estimates of sensitivity and specificity using 95% confidence intervals (CIs).
Main results: We included 57 publications reporting on a total of 54 study cohorts with 15,976 samples, of which 8526 were from cases of SARS-CoV-2 infection. Studies were conducted in Asia (n = 38), Europe (n = 15), and the USA and China (n = 1). We identified data from 25 commercial tests and numerous in-house assays, a small fraction of the 279 antibody assays listed by the Foundation for Innovative Diagnostics. More than half (n = 28) of the studies included were only available as preprints. We had concerns about risk of bias and applicability. Common issues were use of multi-group designs (n = 29), inclusion of only COVID-19 cases (n = 19), lack of blinding of the index test (n = 49) and reference standard (n = 29), differential verification (n = 22), and the lack of clarity about participant numbers, characteristics and study exclusions (n = 47). Most studies (n = 44) only included people hospitalised due to suspected or confirmed COVID-19 infection. There were no studies exclusively in asymptomatic participants. Two-thirds of the studies (n = 33) defined COVID-19 cases based on RT-PCR results alone, ignoring the potential for false-negative RT-PCR results. We observed evidence of selective publication of study findings through omission of the identity of tests (n = 5). We observed substantial heterogeneity in sensitivities of IgA, IgM and IgG antibodies, or combinations thereof, for results aggregated across different time periods post-symptom onset (range 0% to 100% for all target antibodies). We thus based the main results of the review on the 38 studies that stratified results by time since symptom onset. The numbers of individuals contributing data within each study each week are small and are usually not based on tracking the same groups of patients over time. Pooled results for IgG, IgM, IgA, total antibodies and IgG/IgM all showed low sensitivity during the first week since onset of symptoms (all less than 30.1%), rising in the second week and reaching their highest values in the third week. The combination of IgG/IgM had a sensitivity of 30.1% (95% CI 21.4 to 40.7) for 1 to 7 days, 72.2% (95% CI 63.5 to 79.5) for 8 to 14 days, 91.4% (95% CI 87.0 to 94.4) for 15 to 21 days. Estimates of accuracy beyond three weeks are based on smaller sample sizes and fewer studies. For 21 to 35 days, pooled sensitivities for IgG/IgM were 96.0% (95% CI 90.6 to 98.3). There are insufficient studies to estimate sensitivity of tests beyond 35 days post-symptom onset. Summary specificities (provided in 35 studies) exceeded 98% for all target antibodies with confidence intervals no more than 2 percentage points wide. False-positive results were more common where COVID-19 had been suspected and ruled out, but numbers were small and the difference was within the range expected by chance. Assuming a prevalence of 50%, a value considered possible in healthcare workers who have suffered respiratory symptoms, we would anticipate that 43 (28 to 65) would be missed and 7 (3 to 14) would be falsely positive in 1000 people undergoing IgG/IgM testing at days 15 to 21 post-symptom onset. At a prevalence of 20%, a likely value in surveys in high-risk settings, 17 (11 to 26) would be missed per 1000 people tested and 10 (5 to 22) would be falsely positive. At a lower prevalence of 5%, a likely value in national surveys, 4 (3 to 7) would be missed per 1000 tested, and 12 (6 to 27) would be falsely positive. Analyses showed small differences in sensitivity between assay type, but methodological concerns and sparse data prevent comparisons between test brands.
Authors' conclusions: The sensitivity of antibody tests is too low in the first week since symptom onset to have a primary role for the diagnosis of COVID-19, but they may still have a role complementing other testing in individuals presenting later, when RT-PCR tests are negative, or are not done. Antibody tests are likely to have a useful role for detecting previous SARS-CoV-2 infection if used 15 or more days after the onset of symptoms. However, the duration of antibody rises is currently unknown, and we found very little data beyond 35 days post-symptom onset. We are therefore uncertain about the utility of these tests for seroprevalence surveys for public health management purposes. Concerns about high risk of bias and applicability make it likely that the accuracy of tests when used in clinical care will be lower than reported in the included studies. Sensitivity has mainly been evaluated in hospitalised patients, so it is unclear whether the tests are able to detect lower antibody levels likely seen with milder and asymptomatic COVID-19 disease. The design, execution and reporting of studies of the accuracy of COVID-19 tests requires considerable improvement. Studies must report data on sensitivity disaggregated by time since onset of symptoms. COVID-19-positive cases who are RT-PCR-negative should be included as well as those confirmed RT-PCR, in accordance with the World Health Organization (WHO) and China National Health Commission of the People's Republic of China (CDC) case definitions. We were only able to obtain data from a small proportion of available tests, and action is needed to ensure that all results of test evaluations are available in the public domain to prevent selective reporting. This is a fast-moving field and we plan ongoing updates of this living systematic review.
Copyright © 2020 The Authors. Cochrane Database of Systematic Reviews published by John Wiley & Sons, Ltd. on behalf of The Cochrane Collaboration.
Conflict of interest statement
Jonathan J Deeks: none known
Jacqueline Dinnes: none known
Yemisi Takwoingi: none known
Clare Davenport: none known
René Spijker: the Dutch Cochrane Centre (DCC) has received grants for performing commissioned systematic reviews. In no situation, the commissioner had any influence on the results of the work.
Sian Taylor‐Phillips: none known
Ada Adriano: none known
Sophie Beese: none known
Janine Dretzke: none known
Lavinia Ferrante di Ruffano: none known
Isobel Harris: none known
Malcolm Price: none known
Sabine Dittrich: is employed by FIND with funding from DFID and Australian Aid. FIND is a global non‐for profit product development partnership and WHO Diagnostic Collaboration Centre. It is FIND’s role to accelerate access to high quality diagnostic tools for low resource settings and this is achieved by supporting both R&D and access activities for a wide range of diseases, including COVID‐19. .FIND has several clinical research projects to evaluate multiple new diagnostic tests against published Target Product Profiles that have been defined through consensus processes. These studies are for diagnostic products developed by private sector companies who provide access to know‐how, equipment/reagents, and contribute through unrestricted donations as per FIND policy and external SAC review.
Devy Emperador: is employed by FIND with funding from DFID and KFW. FIND is a global non‐for profit product development partnership and WHO Diagnostic Collaboration Centre. It is FIND’s role to accelerate access to high quality diagnostic tools for low resource settings and this is achieved by supporting both R&D and access activities for a wide range of diseases, including COVID‐19. .FIND has several clinical research projects to evaluate multiple new diagnostic tests against published Target Product Profiles that have been defined through consensus processes. These studies are for diagnostic products developed by private sector companies who provide access to know‐how, equipment/reagents, and contribute through unrestricted donations as per FIND policy and external SAC review.
Lotty Hooft: none known
Mariska MG Leeflang: none known
Ann Van den Bruel: none known
Figures



























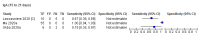

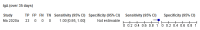

















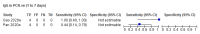




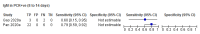
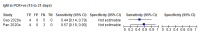














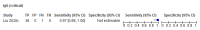
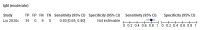


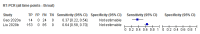


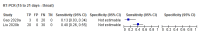



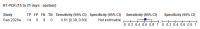
Comment in
-
Antibody tests have higher sensitivity at ≥15 days after symptom onset and 99% specificity for detecting SARS-CoV-2.Ann Intern Med. 2020 Nov 17;173(10):JC57. doi: 10.7326/ACPJ202011170-057. Ann Intern Med. 2020. PMID: 33197346 Free PMC article.
References
References to studies included in this review
Adams 2020 [A] {published data only}
-
- Adams ER, Anand R, Andersson MI, Auckland K, Baillie JK, Barnes E, et al. Evaluation of antibody testing for SARS-Cov-2 using ELISA and lateral flow immunoassays. medRxiv [Preprint] 2020. [www.medrxiv.org/content/10.1101/2020.04.15.20066407v1.full.pdf]
Bendavid 2020 {published data only}
Burbelo 2020 [A] {published data only}
-
- Burbelo PD, Riedo FX, Morishima C, Rawlings S, Smith D, Das S, et al. Detection of nucleocapsid antibody to SARS-CoV-2 is more sensitive than antibody to spike protein in COVID-19 patients. MedRxiv [Preprint] 2020.
Cai 2020a {published data only}
Cassaniti 2020 (A) {published data only}
-
- Cassaniti I, Novazzi F, Giardina F, Salivaro F, Sachs M, Perlini S, et al. Performance of VivaDiagTM COVID-19 IgM/IgG Rapid Test is inadequate for diagnosis of COVID-19 in acute patients referring to emergency room department. Journal of Medical Virology 2020 Mar 30 [Epub ahead of print]. [DOI: 10.1002/jmv.25800] - DOI - PMC - PubMed
Chen 2020a {published data only}
-
- Chen Z, Zhang Z, Zhai X, Li Y, Lin L, Zhao H, et al. Rapid and sensitive detection of anti-SARS-CoV-2 IgG using lanthanide-doped nanoparticles-based lateral flow immunoassay. Analytical Chemistry 2020;92(10):7226-31. - PubMed
Dohla 2020 {published data only}
Du 2020 {published data only}
Freeman 2020 {published data only}
-
- Freeman B, Lester S, Mills L, Rasheed MA, Moye S, Abiona O, et al. Validation of a SARS-CoV-2 spike ELISA for use in contact investigations and serosurveillance. bioRxiv [Preprint] 2020. [DOI: ]
Gao 2020a {published data only}
-
- Gao Y, Yuan Y, Li T T, Wang W X, Li Y X, Li A, et al. Evaluation the auxiliary diagnosis value of antibodies assays for detection of novel coronavirus (SARS-Cov-2) causing an outbreak of pneumonia (COVID-19). medRxiv [Preprint] 2020.
Gao 2020b [A] {published data only}
-
- Gao HX, Li YN, Xu ZG, Wang YL, Wang HB, Cao JF, et al. Detection of serum immunoglobulin M and immunoglobulin G antibodies in 2019-novel coronavirus infected cases from different stages. Chinese Medical Journal (Engl) 2020 Mar 26 [Epub ahead of print]. [DOI: 10.1097/CM9.0000000000000820] - DOI - PMC - PubMed
Garcia 2020 (A) {published data only}
-
- Garcia FP, Perez Tanoira R, Romanyk Cabrera JP, Arroyo Serrano T, Gomez Herruz P, Cuadros Gonzalez J. Rapid diagnosis of SARS-CoV-2 infection by detecting IgG and IgM antibodies with an immunochromatographic device: a prospective single-center study. medRxiv [Preprint] 2020. [DOI: ]
Grzelak 2020 [A] {published data only}
-
- Grzelak L, Temmam S, Planchais C, Demeret C, Huon C, Guivel F, et al. SARS-CoV-2 serological analysis of COVID-19 hospitalized patients, pauci-symptomatic individuals and blood donors. medRxiv [Preprint] 2020. [DOI: ]
Guo 2020a {published data only}
Hu 2020a {published data only}
-
- Hu Q, Cui X, Liu X, Peng B, Jiang J, Wang X, et al. The production of antibodies for SARS-CoV-2 and its clinical implication. medRxiv [Preprint] 2020. [DOI: ]
Infantino 2020 {published data only}
-
- Infantino M, Grossi V, Lari B, Bambi R, Perri A, Manneschi M, et al. Diagnostic accuracy of an automated chemiluminescent immunoassay for anti-SARS-CoV-2 IgM and IgG antibodies: an Italian experience. Journal of Medical Virology 2020 Apr 24 [Epub ahead of print]. [DOI: 10.1002/jmv.25932] - DOI - PMC - PubMed
Jia 2020 {published data only}
Jin 2020 {published data only}
Lassauniere 2020 [A] {published data only}
-
- Lassauniere R, Frische A, Harboe ZB, Nielsen AC, Fomsgaard A, Krogfelt KA, et al. Evaluation of nine commercial SARS-CoV-2 immunoassays. medRxiv [Preprint] 2020. [DOI: ]
Li 2020a {published data only}
Lin 2020a [A] {published data only}
Lippi 2020 [A] {published data only}
-
- Lippi G, Salvagno GL, Pegoraro M, Militello V, Caloi C, Peretti A, et al. Assessment of immune response to SARS-CoV-2 with fully automated MAGLUMI 2019-nCoV IgG and IgM chemiluminescence immunoassays. Clinical Chemistry and Laboratory Medicine 2020 Apr 16 [Epub ahead of print]. [DOI: 10.1515/cclm-2020-0473] - DOI - PubMed
Liu 2020a {published data only}
-
- Liu Y, Liu Y, Diao B, Ren F, Wang Y, Ding J, et al. Diagnostic indexes of a rapid IgG/IgM combined antibody test for SARS-CoV-2. medRxiv [Preprint] 2020. [DOI: ]
Liu 2020b {published data only}
Liu 2020c {published data only}
-
- Liu R, Liu X, Han H, Shereen MA, Niu Z, Li D, et al. The comparative superiority of IgM-IgG antibody test to real-time reverse transcriptase PCR detection for SARS-CoV-2 infection diagnosis. medRxiv [Preprint] 2020. [DOI: ]
Liu 2020d [A] {published data only}
Long 2020 (A) {published data only}
-
- Long Q, Deng H, Chen J, Hu J, Liu B, Liao P, et al. Antibody responses to SARS-CoV-2 in COVID-19 patients: the perspective application of serological tests in clinical practice. medRxiv [Preprint] 2020. [DOI: ]
Lou 2020 [A] {published data only}
Ma 2020a {published data only}
-
- Ma H, Zeng W, He H, Zhao D, Yang Y, Jiang D, et al. COVID-19 diagnosis and study of serum SARS-CoV-2 specific IgA, IgM and IgG by a quantitative and sensitive immunoassay. medRxiv [Preprint] 2020. [DOI: ]
Okba 2020a {published data only}
-
- Woelfel R, Corman VM, Guggemos W, Seilmaier M, Zange S, Muller MA, et al. Virological assessment of hospitalized patients with COVID-2019. Nature 2020;581:465-9. - PubMed
Padoan 2020 {published data only}
-
- Padoan A, Cosma C, Sciacovelli L, Faggian D, Plebani M. Analytical performances of a chemiluminescence immunoassay for SARS-CoV-2 IgM/IgG and antibody kinetics. Clinical Chemistry and Laboratory Medicine 2020. [DOI: doi.org/10.1515/cclm-2020-0443 ] - PubMed
Pan 2020a {published data only}
Paradiso 2020a {published data only}
-
- Paradiso AV, De Summa S, Loconsole D, Procacci V, Sallustio A, Centrone F, et al. Clinical meanings of rapid serological assay in patients tested for SARS-Co2 RT-PCR. medRxiv [Preprint] 2020. [DOI: ]
Qian 2020 {published data only}
-
- Qian C, Zhou M, Cheng F, Lin X, Gong Y, Xie X, et al. Development and multicenter performance evaluation of the first fully automated SARS-CoV-2 IgM and IgG immunoassays. medRxiv [Preprint] 2020. [DOI: ] - PubMed
To 2020a [A] {published data only}
Wan 2020 [A] {published data only}
-
- Wan W Y, Lim S H, Seng E H. Cross-reaction of sera from COVID-19 patients with SARS-CoV assays. medRxiv [Preprint] 2020. [DOI: ] - PubMed
Wang 2020a [A] {published data only}
-
- Wang Q, Du Q, Guo B, Guo Y, Fang L, X Guo. Performance of urea-mediated dissociation in reducing false-positive of 2019-nCoV IgM test. Chinese Journal of Laboratory Medicine 2020. [DOI: 10.3760/cma.j.cn114452-20200219-00091] - DOI
Xiang 2020a [A] {published data only}
-
- Xiang J, Yan M, Li H, Liu T, Lin C, Huang S, et al. Evaluation of enzyme-linked immunoassay and colloidal gold- immunochromatographic assay kit for detection of novel coronavirus (SARS-Cov-2) causing an outbreak of pneumonia (COVID-19). medRxiv [Preprint] 2020. [DOI: ]
Xiang 2020b {published data only}
Xiao 2020a {published data only}
Xie 2020a {published data only}
Xu 2020a {published data only}
-
- Xu Y. Dynamic profile of severe or critical COVID-19 cases. medRxiv [Preprint] 2020. [DOI: ]
Yongchen 2020 {published data only}
Zeng 2020a {published data only}
Zhang 2020a {published data only}
Zhang 2020b {published data only}
Zhang 2020c {published data only}
Zhang 2020d {published data only}
-
- Zhang P, Gao Q, Wang T, Ke Y, Mo F, Jia R, et al. Evaluation of recombinant nucleocapsid and spike proteins for serological diagnosis of novel coronavirus disease 2019 (COVID-19). medRxiv [Preprint] 2020. [DOI: ]
Zhao 2020a {published data only}
Zhao 2020b {published data only}
-
- Zhao R, Li M, Song H, Chen J, Ren W, Feng Y, et al. Serological diagnostic kit of SARS-CoV-2 antibodies using CHO-expressed full-length SARS-CoV-2 S1 proteins. medRxiv [Preprint] 2020. [DOI: ]
References to studies excluded from this review
Ai 2020 {published data only}
Aitken 2020 {published data only}
-
- Aitken J, Ambrose K, Barrell S, Beale R, Bineva-Todd G, Biswas D, et al. Scalable and resilient SARS-CoV2 testing in an academic centre. medRxiv [Preprint] 2020. [DOI: 10.1101/2020.04.19.20071373] - DOI
Amanat 2020 {published data only}
Annamalai 2020 {published data only}
-
- Annamalai P, Kanta M, Ramu P, Ravi B, Veerapandian K, Srinivasan R. A simple colorimetric molecular detection of novel coronavirus (COVID-19), an essential diagnostic tool for pandemic screening. medRxiv [Preprint] 2020. [DOI: 10.1101/2020.04.10.20060293] - DOI
Argenziano 2020 {published data only}
Arons 2020 {published data only}
Arumugam 2020 {published data only}
-
- Arumugam A, Wong SS. The potential use of unprocessed sample for RT-qPCR detection of COVID-19 without an RNA extraction step. bioRxiv [Preprint] 2020. [DOI: 10.1101/2020.04.06.028811] - DOI
Baggett 2020 {published data only}
-
- Baggett TP, Keyes H, Sporn N, Gaeta JM. COVID-19 outbreak at a large homeless shelter in Boston: implications for universal testing. medRxiv [Preprint] 2020. [DOI: 10.1101/2020.04.12.20059618] - DOI
Bai 2020 {published data only}
-
- Bai SL, Wang JY, Zhou YQ, Yu DS, Gao XM, Li LL, et al. Analysis of the first cluster of cases in a family of novel coronavirus pneumonia in Gansu Province. Zhonghua Yu Fang Yi Xue Za Zhi 2020;54(0):E005. [DOI: 10.3760/cma.j.issn.0253-9624.2020.0005 10.3760/cma.j.issn.0253-9624.2020.0005.] - PubMed
Bajema 2020 {published data only}
Barra 2020 {published data only}
Batista 2020 {published data only}
-
- Batista, AF, Miraglia JL, Donato TH, Chiavegatto Filho AD. COVID-19 diagnosis prediction in emergency care patients: a machine learning approach. medRxiv [Preprint] 2020. [DOI: 10.1101/2020.04.04.20052092] - DOI
Beltran Corbellini 2020 {published data only}
-
- Beltran-Corbellini NA, Chico-Garcia JL, Martinez-Poles J, Rodriguez-Jorge F, Natera-Villalba E, Gomez-Corral J, et al. Acute-onset smell and taste disorders in the context of COVID-19: a pilot multicenter PCR-based case-control study. European Journal of Neurology 2020 Apr 22 [Epub ahead of print]. [DOI: 10.1111/ene.14273] - DOI - PMC - PubMed
Beltran Pavez 2020 {published data only}
-
- Beltran-Pavez C, Marquez CL, Munoz G, Valiente-Echeverria F, Gaggero A, Soto-Rifo R, et al. SARS-CoV-2 detection from nasopharyngeal swab samples without RNA extraction. bioRxiv [Preprint] 2020. [DOI: 10.1101/2020.03.28.013508] - DOI
Ben Ami 2020 {published data only}
Bhadra 2020 {published data only}
-
- Bhadra S, Maranhao AC, Ellington AD. A one-enzyme RT-qPCR assay for SARS-CoV-2, and procedures for reagent production. bioRxiv [Preprint] 2020. [DOI: 10.1101/2020.03.29.013342] - DOI
Bordi 2020 {published data only}
-
- Bordi L, Nicastri E, Scorzolini L, Di Caro A, Capobianchi MR, Castilletti C, et al, on behalf of Inmi COVID-Study Group Collaborating Centers. Differential diagnosis of illness in patients under investigation for the novel coronavirus (SARS-CoV-2), Italy, February 2020. Eurosurveillance 2020;25(8):pii=2000170. [DOI: 10.2807/1560-7917.Es.2020.25.8.2000170 10.2807/1560-7917.ES.2020.25.8.2000170] - PMC - PubMed
Burhan 2020 {published data only}
-
- Burhan E, Prasenohadi P, Rogayah R, Isbaniyah F, Reisa T, Dharmawan I. Clinical progression of COVID-19 patient with extended incubation period, delayed RT-PCR time-to-positivity, and potential role of chest CT-scan. Acta Medica Indonesiana 2020;52(1):80-3. - PubMed
Cai 2020 {published data only}
Callahan 2020 {published data only}
Chan 2020 {published data only}
-
- Chan JF, Yuan S, Kok KH, To KK, Chu H, Yang J, et al. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet 2020;395(10223):514-23. [DOI: 10.1016/s0140-6736(20)30154-9 10.1016/S0140-6736(20)30154-9. Epub 2020 Jan 24.] - PMC - PubMed
Chandler Brown 2020 {published data only}
-
- Chandler-Brown D, Bueno AM, Atay O, Tsao DS. A highly scalable and rapidly deployable RNA extraction-free COVID-19 assay by quantitative Sanger sequencing. BioRxiv [Preprint] 2020. [DOI: 10.1101/2020.04.07.029199] - DOI
Chen 2020b {published data only}
-
- Chen X, Ling J, Mo P, Zhang Y, Jiang Q, Ma Z, et al. Restoration of leukomonocyte counts is associated with viral clearance in COVID-19 hospitalized patients. medRxiv [Preprint] 2020. [DOI: 10.1101/2020.03.03.20030437] - DOI
Chen 2020c {published data only}
Cheng 2020a {published data only}
-
- Cheng Z, Lu Y, Cao Q, Qin L, Pan Z, Yan F, et al. Clinical features and chest CT manifestations of coronavirus disease 2019 (COVID-19) in a single-center study in Shanghai, China. American Journal of Roentgenology 2020:1-6. [DOI: 10.2214/ajr.20.22959 10.2214/AJR.20.22959] - PubMed
Chu 2020 {published data only}
Colson 2020 {published data only}
Comar 2020 {published data only}
-
- Comar M, Brumat M, Concas MP, Argentini G, Bianco A, Bicego L, et al. COVID-19 experience: first Italian survey on healthcare staff members from a mother-child research hospital using combined molecular and rapid immunoassays test. MedRxiv [Preprint] 2020. [DOI: 10.1101/2020.04.19.20071563] - DOI
Corman 2020 {published data only}
Cui 2020 {published data only}
-
- Cui P, Chen Z, Wang T, Dai J, Zhang J, Ding T, et al. Clinical features and sexual transmission potential of SARS-CoV-2 infected female patients: a descriptive study in Wuhan, China. medRxiv [preprint] 2020. [DOI: 10.1101/2020.02.26.20028225] - DOI
Curti 2020 {published data only}
-
- Curti L, Pereyra-Bonnet F, Gimenez C. An ultrasensitive, rapid, and portable coronavirus SARS-CoV-2 sequence detection method based on CRISPR-Cas12. bioRxiv [Preprint] 2020. [DOI: 10.1101/2020.02.29.971127] - DOI
Dahlke 2020 {published data only}
-
- Dahlke C, Heidepriem J, Kobbe R, Santer R, Koch T, Fathi A. Distinct early IgA profile may determine severity of COVID-19 symptoms: an immunological case series. medRxiv [Preprint] 2020. [DOI: 10.1101/2020.04.14.20059733] - DOI
Ding 2020 {published data only}
Fang 2020z {published data only}
-
- Fang D, Ma J, Guan J, Wang M, Song Y, Tian D, et al. Manifestations of digestive system in hospitalized patients with novel coronavirus pneumonia in Wuhan, China: a single-center, descriptive study. Chinese Journal of Digestion 2020. [DOI: 10.3760/cma.j.issn. 0254-1432.2020.03.000]
Farfan 2020 {published data only}
Feng 2020 {published data only}
-
- Feng K, Yun YX, Wang XF, Yang GD, Zheng YJ, Lin CM, et al. Analysis of CT features of 15 children with 2019 novel coronavirus infection. Zhonghua Er Ke Za Zhi 2020;58(0):E007. [DOI: 10.3760/cma.j.issn.0578-1310.2020.0007 10.3760/cma.j.issn.0578-1310.2020.0007.] - PubMed
Fontanet 2020 {published data only}
-
- Fontanet A, Tondeur L, Madec Y, Grant R, Besombes C, Jolly N, et al. Cluster of COVID-19 in northern France: a retrospective closed cohort study. medRxiv [Preprint] 2020. [DOI: 10.1101/2020.04.18.20071134] - DOI
Fu 2020a {published data only}
-
- Fu H, Xu H, Zhang N, Xu H, Li Z, Chen H, et al. Association between clinical, laboratory and CT characteristics and RT-PCR results in the follow-up of COVID-19 patients. medRxiv [Preprint] 2020. [DOI: 10.1101/2020.03.19.20038315] - DOI
Fu 2020b {published data only}
Fumeaux 2020 {published data only}
-
- Fumeaux T. COVID-19: Que sait-on des patients admis en soins intensifs? Revue Medicale Suisse 2020;16(690):756. - PubMed
Gao 2020 {published data only}
Giamarellos Bourboulis 2020 {published data only}
Gietema 2020 {published data only}
Gonzalez Gonzalez 2020a {published data only}
-
- Gonzalez-Gonzalez E, Lara-Mayorga IM, Garcia-Rubio A, Garciamendez-Mijares CE, Guerra-Alvarez GE, Garcia-Martinez G, et al. Scaling diagnostics in times of COVID-19: rapid prototyping of 3D-printed water circulators for loop-mediated isothermal amplification (LAMP) and detection of SARS-CoV-2 virus. medRxiv [Preprint] 2020. [DOI: 10.1101/2020.04.09.20058651] - DOI
Gonzalez Gonzalez 2020b {published data only}
-
- Gonzalez-Gonzalez E, Trujillo-de Santiago G, Lara-Mayorga IM, Martinez-Chapa SO, Alvarez MM. Portable and accurate diagnostics for COVID-19: combined use of the miniPCR thermocycler and a well-plate reader for SARS-Co2 virus detection. medRxiv [Preprint] 2020. [DOI: 10.1101/2020.04.03.20052860] - DOI - PMC - PubMed
Guan 2020 {published data only}
-
- Guan WD, Chen LP, Ye F, Ye D, Wu SG, Zhou HX, et al. High-throughput sequencing for confirmation of suspected 2019-nCoV infection identified by fluorescence quantitative polymerase chain reaction. Chinese Medical Journal 2020. [DOI: 10.1097/cm9.0000000000000792 10.1097/CM9.0000000000000792.] - PMC - PubMed
Guo 2020b {published data only}
Guo 2020c {published data only}
-
- Guo X, Guo Z, Duan C, Chen Z, Wang G, Lu Y, et al. Long- term persistence of IgG antibodies in SARS-CoV infected healthcare workers. medRxiv [Preprint] 2020. [DOI: 10.1101/2020.02.12.20021386] - DOI
Han 2020 {published data only}
-
- Han R, Huang L, Jiang H, Dong J, Peng H, Zhang D. Early clinical and CT manifestations of coronavirus disease 2019 (COVID-19) pneumonia. American Journal of Roentgenology 2020:1-6. [DOI: 10.2214/ajr.20.22961 10.2214/AJR.20.22961.] - PubMed
Hao 2020 {published data only}
Hass 2020 {published data only}
Hirotsu 2020 {published data only}
Hogan 2020 {published data only}
Holland 2020 {published data only}
-
- Holland M, Negron D, Mitchell S, Dellinger N, Ivancich M, Barrus T, et al. BioLaboro: a bioinformatics system for detecting molecular assay signature erosion and designing new assays in response to emerging and reemerging pathogens. bioRxiv [Preprint] 2020. [DOI: 10.1101/2020.04.08.031963] - DOI
Hu 2020b {published data only}
-
- Hu F, Chen F, Wang Y, Xu T, Tang X, Li F. Failed detection of the full-length genome of SARS-CoV-2 by ultra-deep sequencing from the recovered and discharged patients retested viral PCR positive. medRxiv [Preprint] 2020. [DOI: 10.1101/2020.03.27.20043299] - DOI
Hu 2020c {published data only}
Hu 2020d {published data only}
Huang 2020a {published data only}
Jenkins 2020 {published data only}
-
- Jenkins C, Orsburn B. In silico approach to accelerate the development of mass spectrometry-based proteomics methods for detection of viral proteins: application to COVID-19. bioRxiv [Preprint] 2020. [DOI: 10.1101/2020.03.08.980383] - DOI
Jiang 2020a {published data only}
Jiang 2020b {published data only}
Jung 2020 {published data only}
-
- Jung YJ, Park GS, Moon JH, Ku K, Beak SH, Kim S, et al. Comparative analysis of primer-probe sets for the laboratory confirmation of SARS-CoV-2. bioRxiv [Preprint] 2020. [DOI: 10.1101/2020.02.25.964775] - DOI
Khan 2020 {published data only}
-
- Khan S, Nakajima R, Jain A, Assis RR, Jasinskas A, Obiero JM, et al. Analysis of serologic cross-reactivity between common human coronaviruses and SARS-CoV-2 using coronavirus antigen microarray. bioRxiv [Preprint] 2020. [DOI: 10.1101/2020.03.24.006544] - DOI
Kim 2019 {published data only}
Kong 2020 {published data only}
Konrad 2020 {published data only}
-
- Konrad R, Eberle U, Dangel A, Treis B, Berger A, Bengs K, et al. Rapid establishment of laboratory diagnostics for the novel coronavirus SARS-CoV-2 in Bavaria, Germany, February 2020. Eurosurveillance 2020;25(9). [DOI: 10.2807/1560-7917.Es.2020.25.9.2000173 10.2807/1560-7917.ES.2020.25.9.2000173.] - PMC - PubMed
Kurstjens 2020 {published data only}
Lamb 2020 {published data only}
-
- Lamb LE, Bartolone SN, Ward E, Chancellor MB. medRxiv [Preprint] 2020. [DOI: 10.1101/2020.02.19.20025155] - DOI
Lan 2020 {published data only}
Lechien 2020 {published data only}
-
- Lechien J, Cabaraux P, Chiesa-Estomba C, Khalife M, Plzak J, Hans S, et al. Objective olfactory testing in patients presenting with sudden onset olfactory dysfunction as the first manifestation of confirmed COVID-19 infection. medRxiv [Preprint] 2020. [DOI: 10.1101/2020.04.15.20066472] - DOI
Lei 2020 {published data only}
Li 2020c {published data only}
Li 2020d {published data only}
-
- Li C, Debruyne D, Spencer J, Kapoor V, Liu LY, Zhang B, et al. High sensitivity detection of SARS-CoV-2 using multiplex PCR and a multiplex-PCR-based metagenomic method. bioRxiv [Preprint] 2020. [DOI: 10.1101/2020.03.12.988246] - DOI
Li 2020e {published data only}
-
- Li Y, Xia L. Coronavirus disease 2019 (COVID-19): role of chest CT in diagnosis and management. American Journal of Roentgenology 2020:1-7. [DOI: 10.2214/ajr.20.22954 10.2214/AJR.20.22954.] - PubMed
Li 2020f {published data only}
-
- Li D, Wang D, Dong J, Wang N, Huang H, Xu H, et al. False- negative results of real-time reverse-transcriptase polymerase chain reaction for severe acute respiratory syndrome coronavirus 2: role of deep-learning-based CT diagnosis and insights from two cases. Korean Journal of Radiology 2020. [DOI: 10.3348/kjr.2020.0146 10.3348/kjr.2020.0146.] - PMC - PubMed
Li 2020g {published data only}
-
- Li YY, Wang WN, Lei Y, Zhang B, Yang J, Hu JW, et al. Comparison of the clinical characteristics between RNA positive and negative patients clinically diagnosed with 2019 novel coronavirus pneumonia. Zhonghua Jie He He Hu Xi Za Zhi 2020;43(0):E023. [DOI: 10.3760/cma.j.cn112147-20200214-00095 10.3760/cma.j.cn112147-20200214-00095.] - PubMed
Liang 2020a {published data only}
-
- Liang Y, Liang J, Zhou Q, Li X, Lin F, Deng Z, et al. Prevalence and clinical features of 2019 novel coronavirus disease (COVID-19) in the fever clinic of a teaching hospital in Beijing: a single-center, retrospective study. medRxiv [Preprint] 2020. [DOI: 10.1101/2020.02.25.20027763] - DOI
Liang 2020b {published data only}
Ling 2020 {published data only}
Liu 2020e {published data only}
Liu 2020f {published data only}
Liu 2020g {published data only}
-
- Liu R, Han H, Liu F, Lv Z, Wu K, Liu Y, et al. Positive rate of RT-PCR detection of SARS-CoV-2 infection in 4880 cases from one hospital in Wuhan, China, from Jan to Feb 2020. Clinica Chimica Acta; International Journal of Clinical Chemistry 2020;505:172-5. [DOI: 10.1016/j.cca.2020.03.009 10.1016/j.cca.2020.03.009.] - PMC - PubMed
Liu 2020h {published data only}
Lo 2020 {published data only}
-
- Lo IL, Lio CF, Cheong HH, Lei CI, Cheong TH, Zhong X, et al. Evaluation of SARS-CoV-2 RNA shedding in clinical specimens and clinical characteristics of 10 patients with COVID-19 in Macau. International Journal of Biological Sciences 2020;16(10):1698-707. [DOI: 10.7150/ijbs.45357] - DOI - PMC - PubMed
Lopez‐Rincon 2020 {published data only}
-
- Lopez-Rincon A, Tonda A, Mendoza-Maldonado L, Claassen E, Garssen J, Kraneveld AD. Accurate identification of SARS-CoV-2 from viral genome sequences using deep learning. bioRxiv [Preprint] 2020. [DOI: 10.1101/2020.03.13.990242] - DOI
Lu 2020 {published data only}
Ma 2020b {published data only}
Mahari 2020 {published data only}
-
- Mahari S, Roberts A, Shahdeo D, Gandhi S. eCovSens-ultrasensitive novel in-house built printed circuit board based electrochemical device for rapid detection of nCOVID-19. bioRxiv [Preprint] 2020. [DOI: 10.1101/2020.04.24.059204] - DOI
Mardani 2020 {published data only}
Marzinotto 2020 {published data only}
McKay 2020 {published data only}
-
- McKay PF, Hu K, Blakney AK, Samnuan K, Bouton CR, Rogers P, et al. Self-amplifying RNA SARS-CoV-2 lipid nanoparticle vaccine induces equivalent preclinical antibody titers and viral neutralization to recovered COVID-19 patients. bioRxiv [Preprint] 2020. [DOI: 10.1101/2020.04.22.055608] - DOI
McRae 2020 {published data only}
Mei 2020 {published data only}
-
- Mei X, Lee HC, Diao K, Huang M, Lin B, Liu C, et al. Artificial intelligence for rapid identification of the coronavirus disease 2019 (COVID-19). medRxiv [Preprint] 2020. [DOI: 10.1101/2020.04.12.20062661] - DOI
Meng 2020 {published data only}
-
- Meng Z, Wang M, Song H, Guo S, Zhou Y, Li W, et al. Development and utilization of an intelligent application for aiding COVID-19 diagnosis. medRxiv [Preprint] 2020. [DOI: 10.1101/2020.03.18.20035816] - DOI
Mercurio 2020 {published data only}
-
- Mercurio I, Tragni V, Busto F, De Grassi A, Pierri CL. Protein structure analysis of the interactions between SARS-CoV-2 spike protein and the human ACE2 receptor: from conformational changes to novel neutralizing antibodies. bioRxiv [Preprint] 2020. [DOI: 10.1101/2020.04.17.046185] - DOI - PMC - PubMed
Metsky 2020 {published data only}
-
- Metsky HC, Freije CA, Kosoko-Thoroddsen TS, Sabeti PC, Myhrvold C. CRISPR-based COVID-19 surveillance using a genomically-comprehensive machine learning approach. bioRxiv [Preprint] 2020. [DOI: 10.1101/2020.02.26.967026] - DOI
Nelson 2020 {published data only}
-
- Nelson AC, Auch B, Schomaker M, Gohl DM, Grady P, Johnson D, et al. Analytical validation of a COVID-19 qRT-PCR detection assay using a 384-well format and three extraction methods. bioRxiv [Preprint] 2020. [DOI: 10.1101/2020.04.02.022186] - DOI
Nemati 2020 {published data only}
-
- Nemati S, Najari HR, Eftekharzadeh A, Kazemifar AM, Qandian A, Fattahi P, et al. Association between rRT-PCR test results upon admission and outcome in hospitalized chest CT-Positive COVID-19 patients; a provincial retrospective cohort with active follow-up. medRxiv [Preprint] 2020. [DOI: 10.1101/2020.04.21.20074641] - DOI
Nie 2020 {published data only}
Nunez Bajo 2020 {published data only}
-
- Nunez-Bajo E, Kasimatis M, Cotur Y, Asfour T, Collins A, Tanriverdi U, et al. Ultra-low-cost integrated silicon-based transducer for on-site, genetic detection of pathogens. bioRxiv [Preprint] 2020. [DOI: 10.1101/2020.03.23.002931] - DOI
Okba 2020b {published data only}
-
- Okba N, Muller MA, Li W, Wang C, GeurtsvanKessel CH, Corman VM, et al. SARS-CoV-2 specific antibody responses in COVID-19 patients. medRxiv [Preprint] 2020. [DOI: 10.1101/2020.03.18.20038059] - DOI
Paden 2020 {published data only}
Pan 2020b {published data only}
Pan 2020c {published data only}
Pan 2020d {published data only}
Pan 2020e {published data only}
Paradiso 2020b {published data only}
-
- Paradiso AV, De Summa S, Silvestris N, Tommasi S, Tufaro A, De Palma G, et al. Rapid serological tests have a role in asymptomatic health workers COVID-19 screening. medRxiv [Preprint] 2020. [DOI: 10.1101/2020.04.15.20057786] - DOI
Park 2020 {published data only}
Peng 2020 {published data only}
Pfefferle 2020 {published data only}
Rauch 2020 {published data only}
Scallan 2020 {published data only}
-
- Scallan MF, Dempsey C, MacSharry J, O'Callaghan I, O'Connor PM, Hogan CP, et al. Validation of a lysis buffer containing 4 M guanidinium thiocyanate (GITC)/ Triton X-100 for extraction of SARS-CoV-2 RNA for COVID-19 testing: comparison of formulated lysis buffers containing 4 to 6 M GITC, Roche external lysis buffer and Qiagen RTL lysis buffer. bioRxiv [Preprint] 2020. [DOI: 10.1101/2020.04.05.026435] - DOI
Seo 2020 {published data only}
Shental 2020 {published data only}
Shi 2020 {published data only}
Shirato 2020 {published data only}
-
- Shirato K, Nao N, Katano H, Takayama I, Saito S, Kato F, et al. Development of genetic diagnostic methods for novel coronavirus 2019 (nCoV-2019) in Japan. Japanese Journal of Infectious Diseases 2020. [DOI: 10.7883/yoken.JJID.2020.061 10.7883/yoken.JJID.2020.061.] - PubMed
Song 2020 {published data only}
-
- Song C, Wang Y, Li W, Hu B, Chen G, Xia P, et al. Detection of 2019 novel coronavirus in semen and testicular biopsy specimen of COVID-19 patients. medRxiv [Preprint] 2020. [DOI: 10.1101/2020.03.31.20042333] - DOI
Su 2020 {published data only}
-
- Su L, Ma X, Yu H, Zhang Z, Bian P, Han Y, et al. The different clinical characteristics of corona virus disease cases between children and their families in China - the character of children with COVID-19. Emerging Microbes and Infections 2020;9(1):707-13. [DOI: 10.1080/22221751.2020.1744483 10.1080/22221751.2020.1744483.] - PMC - PubMed
Sun 2020a {published data only}
Sun 2020b {published data only}
Sun 2020c {published data only}
Tagarro 2020 {published data only}
Tan 2020a {published data only}
-
- Tan W, Lu Y, Zhang J, Wang J, Dan Y, Tan Z, et al. Viral kinetics and antibody responses in patients with COVID-19. medRxiv [Preprint] 2020. [DOI: 10.1101/2020.03.24.20042382] - DOI
Tan 2020b {published data only}
Tan 2020c {published data only}
-
- Tan X, Lin C, Zhang J, Khaing Oo MK, Fan X. Rapid and quantitative detection of COVID-19 markers in micro-liter sized samples. bioRxiv [Preprint] 2020. [DOI: 10.1101/2020.04.20.052233] - DOI
Toptan 2020 {published data only}
Tsang 2003 {published data only}
Vermeiren 2020 {published data only}
Viehweger 2020 {published data only}
-
- Viehweger A, Kühnl F, Brandt C, König B. Increased PCR screening capacity using a multi-replicate pooling scheme. medRxiv [Preprint] 2020. [DOI: 10.1101/2020.04.16.20067603] - DOI
Vogels 2020 {published data only}
Waghmare 2020 {published data only}
Wang 2020b {published data only}
Wang 2020c {published data only}
-
- Wang Z, Li H, Li J, Yang C, Guo X, Hu Z, et al. Elevated serum IgM levels indicate poor outcome in patients with coronavirus disease 2019 pneumonia: a retrospective case-control study. medRxiv [Preprint] 2020. [DOI: 10.1101/2020.03.22.20041285] - DOI
Wang 2020d {published data only}
-
- Wang X, Guo X, Xin Q, Chu Y, Li J, Pan Y, et al. Neutralizing antibodies responses to SARS-CoV-2 in COVID-19 inpatients and convalescent patients. medRxiv [Preprint] 2020. [DOI: 10.1101/2020.04.15.20065623] - DOI
Wang 2020e {published data only}
-
- Wang B, Wang L, Kong X, Geng J, Xiao D, Ma C. Long-term co-existence of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) with antibody response in non-severe coronavirus disease 2019 (COVID-19) patients. medRxiv [Preprint] 2020. [DOI: 10.1101/2020.04.13.20040980] - DOI
Wang 2020f {published data only}
Wang 2020g {published data only}
-
- Wang XF, Yuan J, Zheng YJ, Chen J, Bao YM, Wang YR, et al. Retracted: Clinical and epidemiological characteristics of 34 children with 2019 novel coronavirus infection in Shenzhen. Zhonghua Er Ke Za Zhi 2020;58(0):E008. [DOI: 10.3760/cma.j.issn.0578-1310.2020.0008 10.3760/cma.j.issn.0578-1310.2020.0008.] - PubMed
Wang 2020h {published data only}
-
- Wang QX, Zeng XH, Zheng SL. The nucleic acid test of induced sputum should be used for estimation of patients cure with 2019-nCov. European Review for Medical and Pharmacological Sciences 2020;24(7):3437. [DOI: ] - PubMed
Wang 2020i {published data only}
-
- Wang Z, Weng J, Li Z, Hou R, Zhou L, Ye H, et al. Development and validation of a diagnostic nomogram to predict COVID-19 pneumonia. medRxiv [Preprint] 2020. [DOI: 10.1101/2020.04.03.20052068] - DOI
Wee 2020 {published data only}
Weiss 2020 {published data only}
Woelfel 2020 {published data only}
-
- Woelfel R, Corman VM, Guggemos W, Seilmaier M, Zange S, Mueller MA, et al. Clinical presentation and virological assessment of hospitalized cases of coronavirus disease 2019 in a travel-associated transmission cluster. medRxiv [Preprint] 2020. [DOI: 10.1101/2020.03.05.20030502] - DOI
Won 2020 {published data only}
Woo 2020 {published data only}
-
- Woo CH, Jang S, Shin G, Jung GY, Lee JW. Sensitive one-step isothermal detection of pathogen-derived RNAs. medRxiv [Preprint] 2020. [DOI: 10.1101/2020.03.05.20031971] - DOI
Wu 2020a {published data only}
-
- Wu F, Wang A, Liu M, Wang Q, Chen J, Xia S, et al. Neutralizing antibody responses to SARS-CoV-2 in a COVID-19 recovered patient cohort and their implications. medRxiv [Preprint] 2020. [DOI: 10.1101/2020.03.30.20047365] - DOI
Wu 2020b {published data only}
-
- Wu Q, Xing Y, Shi L, Li W, Gao Y, Pan S, et al. Epidemiological and clinical characteristics of children with coronavirus disease 2019. medRxiv [Preprint] 2020. [DOI: 10.1101/2020.03.19.20027078] - DOI
Wu 2020c {published data only}
Xia 2020a {published data only}
Xia 2020b {published data only}
Xie 2020b {published data only}
Xie 2020c {published data only}
Xing 2020a {published data only}
Xing 2020b {published data only}
-
- Xing Y, Ni W, Wu Q, Li W, Li G, Tong J, et al. Prolonged presence of SARS-CoV-2 in feces of pediatric patients during the convalescent phase. medRxiv [Preprint] 2020. [DOI: 10.1101/2020.03.11.20033159] - DOI
Xu 2020b {published data only}
-
- Xu SP, Kuang D, Hu Y, Liu C, Duan YQ, Wang GP. Detection of 2019-nCoV in the pathological paraffin embedded tissue. Zhonghua Bing Li Xue Za Zhi 2020;49(0):E004. [DOI: 10.3760/cma.j.cn112151.20200219.00001 10.3760/cma.j.cn112151.20200219.00001.] - PubMed
Xu 2020c {published data only}
Xu 2020d {published data only}
Yan 2020 {published data only}
-
- Yan S, Song X, Lin F, Zhu H, Wang X, Li M, et al. Clinical characteristics of coronavirus disease 2019 in Hainan, China. medRxiv [Preprint] 2020. [DOI: 10.1101/2020.03.19.20038539] - DOI
Yang 2020a {published data only}
-
- Yang Y, Yang M, Shen C, Wang F, Yuan J, Li J, et al. Evaluating the accuracy of different respiratory specimens in the laboratory diagnosis and monitoring the viral shedding of 2019-nCoV infections. medRxiv [Preprint] 2020. [DOI: 10.1101/2020.02.11.20021493] - DOI
Yang 2020b {published data only}
Yelin 2020 {published data only}
Yuan 2020 {published data only}
Yun 2020 {published data only}
Zeng 2020b {published data only}
Zhang 2020e {published data only}
-
- Zhang X, Wu X, Wang D, Lu M, Hou X, Wang H, et al. Proteome-wide analysis of differentially-expressed SARS-CoV-2 antibodies in early COVID-19 infection. medRxiv [Preprint] 2020. [DOI: 10.1101/2020.04.14.20064535] - DOI
Zhang 2020f {published data only}
Zhang 2020g {published data only}
Zhang 2020h {published data only}
Zhao 2020c {published data only}
Zhao 2020d {published data only}
-
- Zhao Z, Cui H, Song W, Ru X, Zhou W, Yu X. A simple magnetic nanoparticles-based viral RNA extraction method for efficient detection of SARS-CoV-2. bioRxiv [Preprint] 2020. [DOI: 10.1101/2020.02.22.961268] - DOI
Zhifeng 2020 {published data only}
Zhou 2020 {published data only}
-
- Zhou J, Bai Z, Liu X, Guo Y, Jiang N, Li X, et al. Flocked swab might be one main reason causing the high false-negative rate in COVID-19 screening----the advantages of a novel silicone swab. bioRxiv [Preprint] 2020. [DOI: 10.1101/2020.03.29.014415] - DOI
Zhuang 2020 {published data only}
-
- Zhuang GH, Shen MW, Zeng LX, Mi BB, Chen FY, Liu WJ, et al. Potential false-positive rate among the 'asymptomatic infected individuals' in close contacts of COVID-19 patients. Zhonghua Liu Xing Bing Xue Za Zhi 2020;41(4):485-8. [DOI: 10.3760/cma.j.cn112338-20200221-00144 10.3760/cma.j.cn112338-20200221-00144.] - PubMed
References to studies awaiting assessment
Li 2020b {published data only}
-
- Li H, Li Y, Zhang Z, Lu Z, Wang Y, Lin G, et al. Establishment and clinical performance evaluation of 2019 novel coronavirus antibody colloidal gold detection method. Chinese Journal of Infectious Diseases 2020.
Thompson 2020 {published data only}
Xiong 2020 {published data only}
-
- Xiong Z, Fu L, Zhou H, Liu JK, Wang AM, Huang, Y, et al. Construction and evaluation of a novel diagnosis process for 2019-corona virus disease. Zhonghua Yi Xue Za Zhi 2020;100(0):E019. [DOI: 10.3760/cma.j.cn112137-20200228-00499 10.3760/cma.j.cn112137-20200228-00499.] - PubMed
References to ongoing studies
ChiCTR2000029625 {published data only}
-
- ChiCTR2000029625. Construction of early warning and prediction system for patients with severe / critical novel coronavirus pneumonia (COVID-19). apps.who.int/trialsearch/Trial2.aspx?TrialID=ChiCTR2000029625 (first received 27 April 2020).
ChiCTR2000029695 {published data only}
-
- ChiCTR2000029695. Early detection of novel coronavirus pneumonia (COVID-19) based on a novel high-throughput mass spectrometry analysis with exhaled breath. apps.who.int/trialsearch/Trial2.aspx?TrialID=ChiCTR2000029695 (first received 27 April 2020).
ChiCTR2000029810 {published data only}
-
- ChiCTR2000029810. Clinical study of a novel high sensitivity nucleic acid assay for novel coronavirus pneumonia (COVID-19) based on CRISPR-cas protein. apps.who.int/trialsearch/Trial2.aspx?TrialID=ChiCTR2000029810 (first received 27 April 2020).
ChiCTR2000029870 {published data only}
-
- ChiCTR2000029870. Evaluation of rapid diagnostic kit (IgM/IgG) for novel coronavirus pneumonia (COVID-19). apps.who.int/trialsearch/Trial2.aspx?TrialID=ChiCTR2000029870 (first received 27 April 2020).
ChiCTR2000029883 {published data only}
-
- ChiCTR2000029883. A comparative study on the sensitivity of nasopharyngeal and oropharyngeal swabbing for the detection of SARS-CoV-2 by real-time PCR. apps.who.int/trialsearch/Trial2.aspx?TrialID=ChiCTR2000029883 (first received 27 April 2020).
ChiCTR2000029982 {published data only}
-
- ChiCTR2000029982. Study for using multiomics in the diagnosis and treatment of novel coronavirus pneumonia (COVID-19). apps.who.int/trialsearch/Trial2.aspx?TrialID=ChiCTR2000029982 (first received 27 April 2020).
ChiCTR2000030005 {published data only}
-
- ChiCTR2000030005. Nucleic acid analysis of novel coronavirus pneumonia (COVID-19) in morning sputum samples and pharyngeal swabs-a prospectively diagnostic test. apps.who.int/trialsearch/Trial2.aspx?TrialID=ChiCTR2000030005 (first received 27 April 2020).
ChiCTR2000030085 {published data only}
-
- ChiCTR2000030085. Cancelled by the investigator Study for the false positive rate of IgM / IgG antibody test kit for novel coronavirus pneumonia (COVID-19) in different inpatients. apps.who.int/trialsearch/Trial2.aspx?TrialID=ChiCTR2000030085 (first received 27 April 2020).
ChiCTR2000030185 {published data only}
-
- ChiCTR2000030185. The value of critical care ultrasound in rapid screening, diagnosis, evaluation of effectiveness and intensive prevention of novel coronavirus pneumonia (COVID-19). apps.who.int/trialsearch/Trial2.aspx?TrialID=ChiCTR2000030185 (first received 27 April 2020).
ChiCTR2000030253 {published data only}
-
- ChiCTR2000030253. Exploration and research for a new method for detection of novel coronavirus (COVID-19) nucleic acid. apps.who.int/trialsearch/Trial2.aspx?TrialID=ChiCTR2000030253 (first received 27 April 2020).
ChiCTR2000030334 {published data only}
-
- ChiCTR2000030334. MicroRNA as a marker for early diagnosis of novel coronavirus infection (COVID-19). apps.who.int/trialsearch/Trial2.aspx?TrialID=ChiCTR2000030334 (first received 27 April 2020).
ChiCTR2000030542 {published data only}
-
- ChiCTR2000030542. A clinical study about the diagnosis and prognosis evaluation of novel coronavirus pneumonia (COVID-19) based on viral genome, host genomic sequencing, relative cytokines and other laboratory indexes. apps.who.int/trialsearch/Trial2.aspx?TrialID=ChiCTR2000030542 (first received 27 April 2020).
ChiCTR2000030543 {published data only}
-
- ChiCTR2000030543. Detection of coronavirus in simultaneously collecting tears and throat swab samples collected from the patients with novel coronavirus pneumonia (COVID-19). apps.who.int/trialsearch/Trial2.aspx?TrialID=ChiCTR2000030543 (first received 27 April 2020).
ChiCTR2000030558 {published data only}
-
- ChiCTR2000030558. Cancelled by the investigator Epidemiological research of novel coronavirus pneumonia (COVID-19) suspected cases based on virus nucleic acid test combined with low-dose chest CT screening in primary hospital. apps.who.int/trialsearch/Trial2.aspx?TrialID=ChiCTR2000030558 (first received 27 April 2020).
ChiCTR2000030706 {published data only}
-
- ChiCTR2000030706. Cancelled by the investigator Application of cas13a-mediated RNA detection in the assay of novel coronavirus nucleic acid (COVID-19). apps.who.int/trialsearch/Trial2.aspx?TrialID=ChiCTR2000030706 (first received 27 April 2020).
ChiCTR2000030721 {published and unpublished data}
-
- ChiCTR2000030721. A comparative study for the sensitivity of induced sputum and throat swabs for the detection of SARS-CoV-2 by real-time PCR in patients with novel coronavirus pneumonia (COVID-19). apps.who.int/trialsearch/Trial2.aspx?TrialID=ChiCTR2000030721 (first received 27 April 2020).
ChiCTR2000030754 {published data only}
-
- ChiCTR2000030754. Medical records based study for the accuracy of SARS-CoV-2 IgM antibody screening for diagnosis of novel coronavirus pneumonia (COVID-19). apps.who.int/trialsearch/Trial2.aspx?TrialID=ChiCTR2000030754 (first received 27 April 2020).
ChiCTR2000030833 {published data only}
-
- ChiCTR2000030833. Clinical validation and application of high-throughput novel coronavirus (2019-nCoV) screening detection kit. apps.who.int/trialsearch/Trial2.aspx?TrialID=ChiCTR2000030833 (first received 27 April 2020).
ChiCTR2000030834 {published data only}
-
- ChiCTR2000030834. Epidemiological characteristics and antibody levels of novel coronavirus pneumonia (COVID-19) of pediatric medical staff working in quarantine area. apps.who.int/trialsearch/Trial2.aspx?TrialID=ChiCTR2000030834 (first received 27 April 2020).
ChiCTR2000030838 {published data only}
-
- ChiCTR2000030838. Development of warning system with clinical differential diagnosis and prediction for severe type of novel coronavirus pneumonia (COVID-19) patients based on artificial intelligence and CT images. apps.who.int/trialsearch/Trial2.aspx?TrialID=ChiCTR2000030838 (first received 27 April 2020).
ChiCTR2000030856 {published data only}
-
- ChiCTR2000030856. An artificial intelligence assistant system for suspected novel coronavirus pneumonia (COVID-19) based on chest CT. apps.who.int/trialsearch/Trial2.aspx?TrialID=ChiCTR2000030856 (first received 27 April 2020).
ChiCTR2000030859 {published data only}
-
- ChiCTR2000030859. A medical based analysis for influencing factors of death of novel coronavirus pneumonia (COVID-19) patients in Wuhan Third Hospital. apps.who.int/trialsearch/Trial2.aspx?TrialID=ChiCTR2000030859 (first received 27 April 2020).
ChiCTR2000030860 {published data only}
-
- ChiCTR2000030860. A medical records based study for investigation of dynamic profile of RT-PCR test for SARS-CoV-2 nucleic acid of novel coronavirus pneumonia (COVID-19) patients. apps.who.int/trialsearch/Trial2.aspx?TrialID=ChiCTR2000030860 (first received 27 April 2020).
ChiCTR2000030862 {published data only}
-
- ChiCTR2000030862. Correlation analysis of blood eosinophil cell levels and clinical type category of novel coronavirus pneumonia (COVID-19): a medical records based retrospective study. apps.who.int/trialsearch/Trial2.aspx?TrialID=ChiCTR2000030862 (first received 27 April 2020).
NCT04245631 {published data only}
-
- NCT04245631. Development of a simple, fast and portable recombinase aided amplification assay for 2019-nCoV. clinicaltrials.gov/ct2/show/NCT04245631 (first received 27 April 2020).
NCT04259892 {published data only}
-
- NCT04259892. Viral excretion in contact subjects at high/moderate risk of coronavirus 2019-nCoV infection. ClinicalTrials.gov/show/NCT04259892 (first received 27 April 2020).
NCT04279795 {published data only}
-
- NCT04279795. Detection of 2019 novel coronavirus in multiple organ system and its relationship with clinical manifestations. ClinicalTrials.gov/show/NCT04279795 (first received 27 April 2020).
NCT04304690 {published data only}
-
- NCT04304690. SARS-CoV2 seroconversion among front line medical and paramedical staff in emergency, intensive care units and infectious disease departments during the 2020 epidemic. clinicaltrials.gov/ct2/show/NCT04304690 (first received 27 April 2020).
NCT04311398 {published data only}
-
- NCT04311398. Development and verification of a new coronavirus multiplex nucleic acid detection system. ClinicalTrials.gov/show/NCT04311398 (first received 27 April 2020).
NCT04316728 {published data only}
-
- NCT04316728. Clinical performance of the VivaDiag ™ COVID-19 lgM IgG rapid test in early detecting the infection of COVID-19. ClinicalTrials.gov/show/NCT04316728 (first received 27 April 2020).
NCT04321369 {published data only}
-
- NCT04321369. Impact of swab site and sample collector on testing sensitivity for SARS-CoV-2 virus in symptomatic individuals. clinicaltrials.gov/ct2/show/NCT04321369 (first received 27 April 2020).
NCT04322279 {published data only}
-
- NCT04322279. Factors associated with a positive SARS-CoV-2 serology in contact subjects at high/moderate risk of coronavirus SARS-CoV-2 infection (CoV-CONTACT-SERO). clinicaltrials.gov/ct2/show/NCT04322279 (first received 27 April 2020).
NCT04322513 {published data only}
-
- NCT04322513. Biomarkers for identification of SARS-COV-2 infection. clinicaltrials.gov/ct2/show/NCT04322513 (first received 27 April 2020).
Sullivan 2020 {published data only}
-
- Sullivan PS, Sailey C, Guest JL, Guarner J, Siegler AJ, Valentine-Graves, et al. Detection of SARS-CoV-2 RNA and antibodies in diverse samples: protocol to validate the sufficiency of provider-observed home-collected blood, saliva and oropharyngeal samples. JMIR Public Health and Surveillance 2020. [DOI: 10.2196/19054] - DOI - PMC - PubMed
Additional references
Bossuyt 2015
CDC China 2020
-
- National Health Commission of the People's Republic of China. Release of 7th edition of case definitions. www.nhc.gov.cn/yzygj/s7653p/202003/46c9294a7dfe4cef80dc7f5912eb1989.shtml (accessed prior to 23 June 2020).
Cheng 2020b
Covidence [Computer program]
-
- Veritas Health Innovation Covidence. Version accessed 27 April 2020. Melbourne, Australia: Veritas Health Innovation.Available at covidence.org.
Macaskill 2010
-
- Macaskill P, Gatsonis C, Deeks JJ, Harbord RM, Takwoingi Y. Chapter 10: analysing and presenting results. In: Deeks JJ, Bossuyt PM, Gatsonis C editor(s). Cochrane Handbook for Systematic Reviews of Diagnostic Test Accuracy Version 1.0. The Cochrane Collaboration, 2010. Available from methods.cochrane.org/sdt/handbook-dta-reviews.
McInnes 2018
McInnes 2020
Moher 2009
Review Manager 2014 [Computer program]
-
- Nordic Cochrane Centre, The Cochrane Collaboration Review Manager 5 (RevMan 5). Version 5.3. Copenhagen: Nordic Cochrane Centre, The Cochrane Collaboration, 2014.
Stata [Computer program]
-
- Stata. Version 15. College Station, TX, USA: StataCorp, 2017.Available at www.stata.com.
Takwoingi 2017
Whiting 2011
-
- Whiting PF, Rutjes AW, Westwood ME, Mallett S, Deeks JJ, Reitsma JB, et al. QUADAS‐2: a revised tool for the quality assessment of diagnostic accuracy studies. Annals of Internal Medicine 2011;155(8):529-36. - PubMed
WHO 2020
-
- World Health Organization (WHO). Global surveillance for COVID-19 caused by human infection with COVID-19 virus Interim guidance. www.who.int/publications/i/item/global-surveillance-for-human-infection-... (accessed prior to 23 June 2020). [https://www.who.int/publications/i/item/global-surveillance-for-human-in...]
References to other published versions of this review
Deeks 2020
-
- Deeks JJ, Dinnes J, Takwoingi Y, Davenport C, Leeflang MM, Spijker R, et al. Diagnosis of SARS-CoV-2 infection and COVID-19: accuracy of signs and symptoms; molecular, antigen, and antibody tests; and routine laboratory markers. Cochrane Database of Systematic Reviews 2020, Issue 4. [DOI: 10.1002/14651858.CD013596] - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

