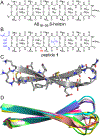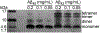X-ray Crystallography Reveals Parallel and Antiparallel β-Sheet Dimers of a β-Hairpin Derived from Aβ16-36 that Assemble to Form Different Tetramers
- PMID: 32584538
- PMCID: PMC7811405
- DOI: 10.1021/acschemneuro.0c00290
X-ray Crystallography Reveals Parallel and Antiparallel β-Sheet Dimers of a β-Hairpin Derived from Aβ16-36 that Assemble to Form Different Tetramers
Abstract
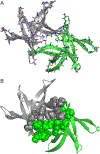
High-resolution structures of oligomers formed by the β-amyloid peptide, Aβ, are important for understanding the molecular basis of Alzheimer's disease. Dimers of Aβ are linked to the pathogenesis and progression of Alzheimer's disease, and tetramers of Aβ are neurotoxic. This paper reports the X-ray crystallographic structures of dimers and tetramers, as well as an octamer, formed by a peptide derived from the central and C-terminal regions of Aβ. In the crystal lattice, the peptide assembles to form two different dimers-an antiparallel β-sheet dimer and a parallel β-sheet dimer-that each further self-assemble to form two different tetramers-a sandwich-like tetramer and a twisted β-sheet tetramer. The structures of these dimers and tetramers derived from Aβ serve as potential models for dimers and tetramers of full-length Aβ that form in vitro and in Alzheimer's disease-afflicted brains.
Keywords: Alzheimer’s disease; Amyloid; Aβ; Crystal structure; Dimer; Oligomer; Tetramer.
Conflict of interest statement
The authors declare no competing financial interest.
Figures