CASC21, a FOXP1 induced long non-coding RNA, promotes colorectal cancer growth by regulating CDK6
- PMID: 32584787
- PMCID: PMC7343488
- DOI: 10.18632/aging.103376
CASC21, a FOXP1 induced long non-coding RNA, promotes colorectal cancer growth by regulating CDK6
Abstract
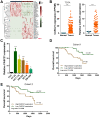
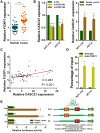
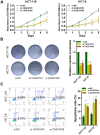
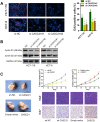
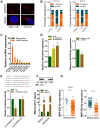
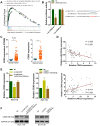
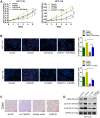
Emerging studies indicate that long non-coding RNAs (lncRNAs) play crucial roles in colorectal cancer (CRC). Here, we reported lncRNA CASC21, which is induced by FOXP1, functions as an oncogene in CRC. We systematically elucidated its clinical significance and possible molecular mechanism in CRC. LncRNA expression in CRC was analyzed by RNA-sequencing data in TCGA. The expression level of CASC21 in tissues was determined by qRT-PCR. The functions of CASC21 was investigated by in vitro and in vivo assays (CCK8 assay, colony formation assay, EdU assay, xenograft model, flow cytometry assay, immunohistochemistry (IHC) and Western blot). Chromatin immunoprecipitation (ChIP), RNA immunoprecipitation (RIP) and luciferase reporter assays were utilized to demonstrate the potential mechanisms of CASC21. CASC21 is overexpressed in CRC and high CASC21 expression is associated with poor survival. Functional experiments revealed that CASC21 promotes CRC cell growth. Mechanistically, we found that CASC21 expressed predominantly in the cytoplasm. CASC21 could interact with miR-539-5p and regulate its target CDK6. Together, our study elucidated that CASC21 acted as an oncogene in CRC, which might serve as a novel target for CRC diagnosis and therapy.
Keywords: CASC21; CDK6; colorectal cancer; lncRNA; miR-539-5p.
Conflict of interest statement
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

