Neutralization of SARS-CoV-2 by Destruction of the Prefusion Spike
- PMID: 32585135
- PMCID: PMC7303615
- DOI: 10.1016/j.chom.2020.06.010
Neutralization of SARS-CoV-2 by Destruction of the Prefusion Spike
Erratum in
-
Neutralization of SARS-CoV-2 by Destruction of the Prefusion Spike.Cell Host Microbe. 2020 Sep 9;28(3):497. doi: 10.1016/j.chom.2020.07.002. Cell Host Microbe. 2020. PMID: 32910920 Free PMC article. No abstract available.
Abstract
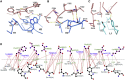
There are as yet no licensed therapeutics for the COVID-19 pandemic. The causal coronavirus (SARS-CoV-2) binds host cells via a trimeric spike whose receptor binding domain (RBD) recognizes angiotensin-converting enzyme 2, initiating conformational changes that drive membrane fusion. We find that the monoclonal antibody CR3022 binds the RBD tightly, neutralizing SARS-CoV-2, and report the crystal structure at 2.4 Å of the Fab/RBD complex. Some crystals are suitable for screening for entry-blocking inhibitors. The highly conserved, structure-stabilizing CR3022 epitope is inaccessible in the prefusion spike, suggesting that CR3022 binding facilitates conversion to the fusion-incompetent post-fusion state. Cryogenic electron microscopy (cryo-EM) analysis confirms that incubation of spike with CR3022 Fab leads to destruction of the prefusion trimer. Presentation of this cryptic epitope in an RBD-based vaccine might advantageously focus immune responses. Binders at this epitope could be useful therapeutically, possibly in synergy with an antibody that blocks receptor attachment.
Keywords: CR3022; SARS-CoV-2; X-ray crystallography; antibody; cryo-electron microscopy; epitope; neutralization; receptor binding domain; spike; therapeutic.
Copyright © 2020 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Interests The authors declare no competing interests.
Figures







References
-
- Aricescu A.R., Lu W., Jones E.Y. A time- and cost-efficient system for high-level protein production in mammalian cells. Acta Crystallogr. D Biol. Crystallogr. 2006;62:1243–1250. - PubMed
-
- Emsley P., Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr. Sect. D Biol. Crystallogr. 2004;60:2126–2132. - PubMed
-
- Grist N.R. Oxford, Blackwell Scientific; 1966. Diagnostic methods in clinical virology.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

