Genetic Diversity of Potassium Ion Channel Proteins Encoded by Chloroviruses That Infect Chlorella heliozoae
- PMID: 32585987
- PMCID: PMC7354518
- DOI: 10.3390/v12060678
Genetic Diversity of Potassium Ion Channel Proteins Encoded by Chloroviruses That Infect Chlorella heliozoae
Abstract
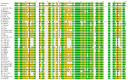
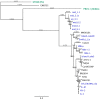
Chloroviruses are large, plaque-forming, dsDNA viruses that infect chlorella-like green algae that live in a symbiotic relationship with protists. Chloroviruses have genomes from 290 to 370 kb, and they encode as many as 400 proteins. One interesting feature of chloroviruses is that they encode a potassium ion (K+) channel protein named Kcv. The Kcv protein encoded by SAG chlorovirus ATCV-1 is one of the smallest known functional K+ channel proteins consisting of 82 amino acids. The KcvATCV-1 protein has similarities to the family of two transmembrane domain K+ channel proteins; it consists of two transmembrane α-helixes with a pore region in the middle, making it an ideal model for studying K+ channels. To assess their genetic diversity, kcv genes were sequenced from 103 geographically distinct SAG chlorovirus isolates. Of the 103 kcv genes, there were 42 unique DNA sequences that translated into 26 new Kcv channels. The new predicted Kcv proteins differed from KcvATCV-1 by 1 to 55 amino acids. The most conserved region of the Kcv protein was the filter, the turret and the pore helix were fairly well conserved, and the outer and the inner transmembrane domains of the protein were the most variable. Two of the new predicted channels were shown to be functional K+ channels.
Keywords: Chloroviruses; Kcv channels; algal viruses; potassium ion channels.
Conflict of interest statement
The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures





References
-
- Cherrier M.V., Kostyuchenko V.A., Xiao C., Bowman V.D., Battisti A.J., Yan X., Chipman P.R., Baker T.S., Van Etten J.L., Rossmann M.G. An icosahedral algal virus has a complex unique vertex decorated by a spike. Proc. Natl. Acad. Sci. USA. 2009;106:11085–11089. doi: 10.1073/pnas.0904716106. - DOI - PMC - PubMed
-
- Milrot E., Shimoni E., Dadosh T., Rechav K., Unger T., Van Etten J.L., Minsky A. Structural studies demonstrating a bacteriophage-like replication cycle of the eukaryote-infecting Paramecium bursaria chlorella virus-1. PLoS Pathog. 2017;13:e1006562. doi: 10.1371/journal.ppat.1006562. - DOI - PMC - PubMed
-
- Romani G., Piotrowski A., Hillmer S., Gurnon J., Van Etten J.L., Moroni A., Thiel G., Hertel B. A virus-encoded potassium ion channel is a structural protein in the chlorovirus Paramecium bursaria chlorella virus 1 virion. J. Gen. Virol. 2013;94:2549–2556. doi: 10.1099/vir.0.055251-0. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

