The OsGSK2 Kinase Integrates Brassinosteroid and Jasmonic Acid Signaling by Interacting with OsJAZ4
- PMID: 32586913
- PMCID: PMC7474301
- DOI: 10.1105/tpc.19.00499
The OsGSK2 Kinase Integrates Brassinosteroid and Jasmonic Acid Signaling by Interacting with OsJAZ4
Abstract
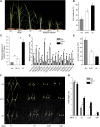
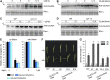
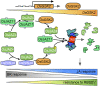
The crosstalk between brassinosteroid (BR) and jasmonic acid (JA) signaling is crucial for plant growth and defense responses. However, the detailed interplay between BRs and JA remains obscure. Here, we found that the rice (Oryza sativa) Glycogen synthase kinase3 (GSK3)-like kinase OsGSK2, a conserved kinase serving as a key suppressor of BR signaling, enhanced antiviral defense and the JA response. We identified a member of the JASMONATE ZIM-domain (JAZ) family, OsJAZ4, as a OsGSK2 substrate and confirmed that OsGSK2 interacted with and phosphorylated OsJAZ4. We demonstrated that OsGSK2 disrupted the OsJAZ4-OsNINJA complex and OsJAZ4-OsJAZ11 dimerization by competitively binding to the ZIM domain, perhaps helping to facilitate the degradation of OsJAZ4 via the 26S proteasome pathway. We also showed that OsJAZ4 negatively modulated JA signaling and antiviral defense and that the BR pathway was involved in modulating the stability of OsJAZ4 protein in an OsCORONATINE INSENSITIVE1-dependent manner. Collectively, these results suggest that OsGSK2 enhances plant antiviral defenses by activating JA signaling as it directly interacts with, phosphorylates, and destabilizes OsJAZ4. Thus, our findings provide a clear link between BR and JA signaling.
© 2020 American Society of Plant Biologists. All rights reserved.
Figures






Comment in
-
OsGSK2 Integrates Jasmonic Acid and Brassinosteroid Signaling in Rice.Plant Cell. 2020 Sep;32(9):2669-2670. doi: 10.1105/tpc.20.00531. Epub 2020 Jul 14. Plant Cell. 2020. PMID: 32665308 Free PMC article. No abstract available.
References
-
- Cai Z., Liu J., Wang H., Yang C., Chen Y., Li Y., Pan S., Dong R., Tang G., Barajas-Lopez JDE.D., Fujii H., Wang X.(2014). GSK3-like kinases positively modulate abscisic acid signaling through phosphorylating subgroup III SnRK2s in Arabidopsis. Proc. Natl. Acad. Sci. USA 111: 9651–9656. - PMC - PubMed
-
- Chini A., Fonseca S., Chico J.M., Fernández-Calvo P., Solano R.(2009). The ZIM domain mediates homo- and heteromeric interactions between Arabidopsis JAZ proteins. Plant J. 59: 77–87. - PubMed
-
- Chini A., Fonseca S., Fernández G., Adie B., Chico J.M., Lorenzo O., García-Casado G., López-Vidriero I., Lozano F.M., Ponce M.R., Micol J.L., Solano R.(2007). The JAZ family of repressors is the missing link in jasmonate signalling. Nature 448: 666–671. - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources

