Small Molecule Inhibitors Confirm Ubiquitin-Dependent Removal of TOP2-DNA Covalent Complexes
- PMID: 32587095
- PMCID: PMC7416847
- DOI: 10.1124/mol.119.118893
Small Molecule Inhibitors Confirm Ubiquitin-Dependent Removal of TOP2-DNA Covalent Complexes
Abstract
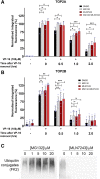
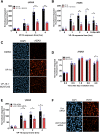
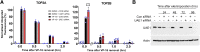
DNA topoisomerase II (TOP2) is required for the unwinding and decatenation of DNA through the induction of an enzyme-linked double-strand break (DSB) in one DNA molecule and passage of another intact DNA duplex through the break. Anticancer drugs targeting TOP2 (TOP2 poisons) prevent religation of the DSB and stabilize a normally transient intermediate of the TOP2 reaction mechanism called the TOP2-DNA covalent complex. Subsequently, TOP2 remains covalently bound to each end of the enzyme-bridged DSB, which cannot be repaired until TOP2 is removed from the DNA. One removal mechanism involves the proteasomal degradation of the TOP2 protein, leading to the liberation of a protein-free DSB. Proteasomal degradation is often regulated by protein ubiquitination, and here we show that inhibition of ubiquitin-activating enzymes reduces the processing of TOP2A- and TOP2B-DNA complexes. Depletion or inhibition of ubiquitin-activating enzymes indicated that ubiquitination was required for the liberation of etoposide-induced protein-free DSBs and is therefore an important layer of regulation in the repair of TOP2 poison-induced DNA damage. TOP2-DNA complexes stabilized by etoposide were shown to be conjugated to ubiquitin, and this was reduced by inhibition or depletion of ubiquitin-activating enzymes. SIGNIFICANCE STATEMENT: There is currently great clinical interest in the ubiquitin-proteasome system and ongoing development of specific inhibitors. The results in this paper show that the therapeutic cytotoxicity of DNA topoisomerase II (TOP2) poisons can be enhanced through combination therapy with ubiquitin-activating enzyme inhibitors or by specific inhibition of the BMI/RING1A ubiquitin ligase, which would lead to increased cellular accumulation or persistence of TOP2-DNA complexes.
Copyright © 2020 The Author(s).
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures



Similar articles
-
A Role for VCP/p97 in the Processing of Drug-Stabilized TOP2-DNA Covalent Complexes.Mol Pharmacol. 2021 Jul;100(1):57-62. doi: 10.1124/molpharm.121.000262. Epub 2021 May 3. Mol Pharmacol. 2021. PMID: 33941661 Free PMC article.
-
Suppressing proteasome mediated processing of topoisomerase II DNA-protein complexes preserves genome integrity.Elife. 2020 Feb 14;9:e53447. doi: 10.7554/eLife.53447. Elife. 2020. PMID: 32057297 Free PMC article.
-
Effect of TDP2 on the Level of TOP2-DNA Complexes and SUMOylated TOP2-DNA Complexes.Int J Mol Sci. 2018 Jul 14;19(7):2056. doi: 10.3390/ijms19072056. Int J Mol Sci. 2018. PMID: 30011940 Free PMC article.
-
Regulation of topoisomerase II stability and activity by ubiquitination and SUMOylation: clinical implications for cancer chemotherapy.Mol Biol Rep. 2021 Sep;48(9):6589-6601. doi: 10.1007/s11033-021-06665-7. Epub 2021 Sep 2. Mol Biol Rep. 2021. PMID: 34476738 Free PMC article. Review.
-
Mechanisms to Repair Stalled Topoisomerase II-DNA Covalent Complexes.Mol Pharmacol. 2022 Jan;101(1):24-32. doi: 10.1124/molpharm.121.000374. Epub 2021 Oct 23. Mol Pharmacol. 2022. PMID: 34689119 Review.
Cited by
-
Identification of the Potential Prognosis Biomarkers in Hepatocellular Carcinoma: An Analysis Based on WGCNA and PPI.Int J Gen Med. 2021 Dec 10;14:9555-9565. doi: 10.2147/IJGM.S338500. eCollection 2021. Int J Gen Med. 2021. PMID: 34916837 Free PMC article.
-
BRCA1-BARD1 regulates transcription through modulating topoisomerase IIβ.Open Biol. 2021 Oct;11(10):210221. doi: 10.1098/rsob.210221. Epub 2021 Oct 6. Open Biol. 2021. PMID: 34610268 Free PMC article.
-
Expression and biological significance of topoisomerase II α (TOP2A) in oral squamous cell carcinoma.Discov Oncol. 2024 Sep 10;15(1):423. doi: 10.1007/s12672-024-01295-4. Discov Oncol. 2024. PMID: 39254737 Free PMC article.
-
Investigating Transcriptional Dynamics Changes and Time-Dependent Marker Gene Expression in the Early Period After Skeletal Muscle Injury in Rats.Front Genet. 2021 Jun 17;12:650874. doi: 10.3389/fgene.2021.650874. eCollection 2021. Front Genet. 2021. PMID: 34220936 Free PMC article.
-
The abscission checkpoint senses chromatin bridges through Top2α recruitment to DNA knots.J Cell Biol. 2023 Nov 6;222(11):e202303123. doi: 10.1083/jcb.202303123. Epub 2023 Aug 28. J Cell Biol. 2023. PMID: 37638884 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

