Loss of Complement Factor H impairs antioxidant capacity and energy metabolism of human RPE cells
- PMID: 32587311
- PMCID: PMC7316856
- DOI: 10.1038/s41598-020-67292-z
Loss of Complement Factor H impairs antioxidant capacity and energy metabolism of human RPE cells
Abstract
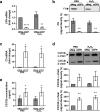
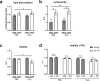
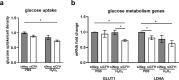
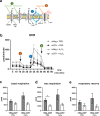
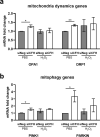
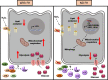
Polymorphisms in the Complement Factor H (CFH) gene, coding for the Factor H protein (FH), can increase the risk for age-related macular degeneration (AMD). AMD-associated CFH risk variants, Y402H in particular, impair FH function leading to complement overactivation. Whether this alone suffices to trigger AMD pathogenesis remains unclear. In AMD, retinal homeostasis is compromised due to the dysfunction of retinal pigment epithelium (RPE) cells. To investigate the impact of endogenous FH loss on RPE cell balance, we silenced CFH in human hTERT-RPE1 cells. FH reduction led to accumulation of C3, at both RNA and protein level and increased RPE vulnerability toward oxidative stress. Mild hydrogen-peroxide exposure in combination with CFH knock-down led to a reduction of glycolysis and mitochondrial respiration, paralleled by an increase in lipid peroxidation, which is a key aspect of AMD pathogenesis. In parallel, cell viability was decreased. The perturbations of energy metabolism were accompanied by transcriptional deregulation of several glucose metabolism genes as well as genes modulating mitochondrial stability. Our data suggest that endogenously produced FH contributes to transcriptional and metabolic homeostasis and protects RPE cells from oxidative stress, highlighting a novel role of FH in AMD pathogenesis.
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
Complement Factor H Loss in RPE Cells Causes Retinal Degeneration in a Human RPE-Porcine Retinal Explant Co-Culture Model.Biomolecules. 2021 Nov 3;11(11):1621. doi: 10.3390/biom11111621. Biomolecules. 2021. PMID: 34827622 Free PMC article.
-
CFH Loss in Human RPE Cells Leads to Inflammation and Complement System Dysregulation via the NF-κB Pathway.Int J Mol Sci. 2021 Aug 13;22(16):8727. doi: 10.3390/ijms22168727. Int J Mol Sci. 2021. PMID: 34445430 Free PMC article.
-
Impaired Mitochondrial Function in iPSC-Retinal Pigment Epithelium with the Complement Factor H Polymorphism for Age-Related Macular Degeneration.Cells. 2021 Apr 2;10(4):789. doi: 10.3390/cells10040789. Cells. 2021. PMID: 33918210 Free PMC article.
-
NAD+ inhibits the metabolic reprogramming of RPE cells in early AMD by upregulating mitophagy.Discov Med. 2019 May;27(149):189-196. Discov Med. 2019. PMID: 31361981 Review.
-
Retinal pigment epithelium in the pathogenesis of age-related macular degeneration and photobiomodulation as a potential therapy?Clin Exp Ophthalmol. 2018 Aug;46(6):670-686. doi: 10.1111/ceo.13121. Epub 2018 Jan 12. Clin Exp Ophthalmol. 2018. PMID: 29205705 Review.
Cited by
-
Complement Factor H Loss in RPE Cells Causes Retinal Degeneration in a Human RPE-Porcine Retinal Explant Co-Culture Model.Biomolecules. 2021 Nov 3;11(11):1621. doi: 10.3390/biom11111621. Biomolecules. 2021. PMID: 34827622 Free PMC article.
-
Effects of E-Cigarette Refill Liquid Flavorings with and without Nicotine on Human Retinal Pigment Epithelial Cells: A Preliminary Study.Int J Environ Res Public Health. 2021 Nov 6;18(21):11655. doi: 10.3390/ijerph182111655. Int J Environ Res Public Health. 2021. PMID: 34770169 Free PMC article.
-
Organ Cultures for Retinal Diseases.Front Neurosci. 2020 Nov 25;14:583392. doi: 10.3389/fnins.2020.583392. eCollection 2020. Front Neurosci. 2020. PMID: 33324149 Free PMC article. Review.
-
RPE-Directed Gene Therapy Improves Mitochondrial Function in Murine Dry AMD Models.Int J Mol Sci. 2023 Feb 14;24(4):3847. doi: 10.3390/ijms24043847. Int J Mol Sci. 2023. PMID: 36835257 Free PMC article.
-
The AMD-associated genetic polymorphism CFH Y402H confers vulnerability to Hydroquinone-induced stress in iPSC-RPE cells.Front Immunol. 2025 Feb 6;16:1527018. doi: 10.3389/fimmu.2025.1527018. eCollection 2025. Front Immunol. 2025. PMID: 39981241 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

