Genetic Organization of the aprX-lipA2 Operon Affects the Proteolytic Potential of Pseudomonas Species in Milk
- PMID: 32587583
- PMCID: PMC7298200
- DOI: 10.3389/fmicb.2020.01190
Genetic Organization of the aprX-lipA2 Operon Affects the Proteolytic Potential of Pseudomonas Species in Milk
Abstract
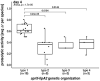
Psychrotolerant Pseudomonas species are a main cause of proteolytic spoilage of ultra-high temperature (UHT) milk products due to the secretion of the heat-resistant metallopeptidase AprX, which is encoded by the first gene of the aprX-lipA2 operon. While the proteolytic property has been characterized for many different Pseudomonas isolates, the underlying aprX-lipA2 gene organization was only described for a few strains so far. In this study, the phylogenomic analysis of 185 Pseudomonas type strains revealed that the presence of aprX is strongly associated to a monophylum composed of 81 species, of which 83% carried the aprX locus. Furthermore, almost all type strains of known milk-relevant species were shown to be members of the three monophyletic groups P. fluorescens, P. gessardii, and P. fragi. In total, 22 different types of aprX-lipA2 genetic organizations were identified in the genus, whereby 31% of the species tested carried the type 1 operon structure consisting of eight genes (aprXIDEF prtAB lipA2). Other genetic structures differed from type 1 mainly in the presence and location of genes coding for two lipases (lipA1 and lipA2) and putative autotransporters (prtA and prtB). The peptidase activity of 129 strains, as determined on skim milk agar and in UHT-milk, correlated largely with different aprX-lipA2 gene compositions. Particularly, isolates harboring the type 1 operon were highly proteolytic, while strains with other operon types, especially ones lacking prtA and prtB, exhibited significantly lower peptidase activities. In conclusion, the phylogenomic position and the aprX-lipA2 gene organization specify the proteolytic potential of Pseudomonas isolates. In addition, however, an interplay of several environmental factors and intrinsic traits influences production and activity of AprX, leading to strain-specific proteolytic phenotypes.
Keywords: AprX peptidase; aprX-lipA2 operon; genus Pseudomonas; milk spoilage; proteolytic potential.
Copyright © 2020 Maier, Huptas, von Neubeck, Scherer, Wenning and Lücking.
Figures




References
-
- Arnold J. B. (2019). ggthemes: extra themes, scales and geoms for ‘ggplot2’ [Online]. Available at: https://CRAN.R-project.org/package=ggthemes (Accessed March 19, 2020).
-
- Auguie B. (2017). gridExtra: miscellaneous functions for "Grid" graphics [Online]. Available at: https://CRAN.R-project.org/package=gridExtra (Accessed March 19, 2020).
-
- Baur C., Krewinkel M., Kutzli I., Kranz B., von Neubeck M., Huptas C., et al. (2015b). Isolation and characterisation of a heat-resistant peptidase from Pseudomonas panacis withstanding general UHT processes. Int. Dairy J. 49, 46–55. 10.1016/j.idairyj.2015.04.009 - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources

