In vivo Screen Identifies Zdhhc2 as a Critical Regulator of Germinal Center B Cell Differentiation
- PMID: 32587588
- PMCID: PMC7297983
- DOI: 10.3389/fimmu.2020.01025
In vivo Screen Identifies Zdhhc2 as a Critical Regulator of Germinal Center B Cell Differentiation
Abstract
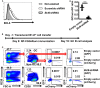
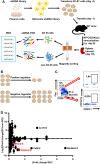
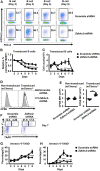
Germinal center (GC) B cell differentiation is critical for the production of affinity-matured pathogen-specific antibodies, the dysregulation of which may lead to humoral immunodeficiency or autoimmunity. The development of an in vivo screening system for factors regulating GC B cell differentiation has been a challenge. Here we describe a small-scale in vivo screening system with NP-specific B1-8hi cells and a retroviral shRNA library targeting 78 candidate genes to search for B cell-intrinsic factors that specifically regulate GC B cell differentiation. Zdhhc2, a gene encoding palmitoyltransferase ZDHHC2 and highly expressed in GC B cells, is identified as a strong positive regulator of GC B cell differentiation. B1-8hi cells transduced with Zdhhc2-shRNA are severely compromised in differentiating into GC B cells. A further analysis of in vitro differentiated B cells transduced with Zdhhc2-shRNA shows that Zdhhc2 is critical for the proliferation and the survival of B cells stimulated by CD40L, BAFF, and IL-21 and consequently impacts on their differentiation into GC B cells and post-GC B cells. These studies not only identify Zdhhc2 as a novel regulator of GC B cell differentiation but also represent a proof of concept of in vivo screen for regulators of GC B cell differentiation.
Keywords: GC selection; Zdhhc2; germinal center B cell; in vivo screen; shRNA.
Copyright © 2020 Zhao, Zhang, Zhang, Li, Huang and Li.
Figures



Similar articles
-
BAFF, IL-4 and IL-21 separably program germinal center-like phenotype acquisition, BCL6 expression, proliferation and survival of CD40L-activated B cells in vitro.Immunol Cell Biol. 2019 Oct;97(9):826-839. doi: 10.1111/imcb.12283. Epub 2019 Jul 29. Immunol Cell Biol. 2019. PMID: 31276232
-
T Follicular Helper Cell-Germinal Center B Cell Interaction Strength Regulates Entry into Plasma Cell or Recycling Germinal Center Cell Fate.Immunity. 2018 Apr 17;48(4):702-715.e4. doi: 10.1016/j.immuni.2018.03.027. Immunity. 2018. PMID: 29669250
-
Augmented antibody response with premature germinal center regression in CD40L transgenic mice.J Immunol. 2010 Jul 1;185(1):211-9. doi: 10.4049/jimmunol.0901694. Epub 2010 May 26. J Immunol. 2010. PMID: 20505144
-
The primary germinal center response in mice.Curr Opin Immunol. 2005 Jun;17(3):298-302. doi: 10.1016/j.coi.2005.04.007. Curr Opin Immunol. 2005. PMID: 15886120 Review.
-
Regulation of the Germinal Center Response.Front Immunol. 2018 Oct 25;9:2469. doi: 10.3389/fimmu.2018.02469. eCollection 2018. Front Immunol. 2018. PMID: 30410492 Free PMC article. Review.
Cited by
-
Forward and Reverse Genetics of B Cell Malignancies: From Insertional Mutagenesis to CRISPR-Cas.Front Immunol. 2021 Aug 13;12:670280. doi: 10.3389/fimmu.2021.670280. eCollection 2021. Front Immunol. 2021. PMID: 34484175 Free PMC article. Review.
-
Shared Pathogenicity Features and Sequences between EBV, SARS-CoV-2, and HLA Class I Molecule-binding Motifs with a Potential Role in Autoimmunity.Clin Rev Allergy Immunol. 2023 Aug;65(2):206-230. doi: 10.1007/s12016-023-08962-4. Epub 2023 Jul 28. Clin Rev Allergy Immunol. 2023. PMID: 37505416 Review.
-
ZDHHC2 promoted antimycobacterial responses by selective autophagic degradation of B-RAF and C-RAF in macrophages.Sci Adv. 2025 Jan 24;11(4):eadq7706. doi: 10.1126/sciadv.adq7706. Epub 2025 Jan 24. Sci Adv. 2025. PMID: 39854453 Free PMC article.
-
Protein posttranslational modifications in health and diseases: Functions, regulatory mechanisms, and therapeutic implications.MedComm (2020). 2023 May 2;4(3):e261. doi: 10.1002/mco2.261. eCollection 2023 Jun. MedComm (2020). 2023. PMID: 37143582 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

