Recommendations for Choosing the Genotyping Method and Best Practices for Quality Control in Crop Genome-Wide Association Studies
- PMID: 32587600
- PMCID: PMC7299185
- DOI: 10.3389/fgene.2020.00447
Recommendations for Choosing the Genotyping Method and Best Practices for Quality Control in Crop Genome-Wide Association Studies
Abstract
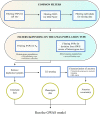
High-throughput genotyping boosts genome-wide association studies (GWAS) in crop species, leading to the identification of single-nucleotide polymorphisms (SNPs) associated with economically important traits. Choosing a cost-effective genotyping method for crop GWAS requires careful examination of several aspects, namely, the purpose and the scale of the study, crop-specific genomic features, and technical and economic matters associated with each genotyping option. Once genotypic data have been obtained, quality control (QC) procedures must be applied to avoid bias and false signals in genotype-phenotype association tests. QC for human GWAS has been extensively reviewed; however, QC for crop GWAS may require different actions, depending on the GWAS population type. Here, we review most popular genotyping methods based on next-generation sequencing (NGS) and array hybridization, and report observations that should guide the investigator in the choice of the genotyping method for crop GWAS. We provide recommendations to perform QC in crop species, and deliver an overview of bioinformatics tools that can be used to accomplish all needed tasks. Overall, this work aims to provide guidelines to harmonize those procedures leading to SNP datasets ready for crop GWAS.
Keywords: GWAS; bioinformatics tools; crops; genotyping; quality control.
Copyright © 2020 Pavan, Delvento, Ricciardi, Lotti, Ciani and D’Agostino.
Figures
References
-
- Allen A. M., Winfield M. O., Burridge A. J., Downie R. C., Benbow H. R., Barker G. L. A., et al. (2017). Characterization of a wheat breeders’ array suitable for high-throughput SNP genotyping of global accessions of hexaploid bread wheat (Triticum aestivum). Plant Biotechnol. J. 15 390–401. 10.1111/pbi.12635 - DOI - PMC - PubMed
-
- Astle W., Balding D. J. (2009). Population structure and cryptic relatedness in genetic association studies. Stat. Sci. 24 451–471. 10.1214/09-STS307 - DOI
Publication types
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

