Phosphodiesterase 10A Is a Mediator of Osteogenic Differentiation and Mechanotransduction in Bone Marrow-Derived Mesenchymal Stromal Cells
- PMID: 32587621
- PMCID: PMC7294361
- DOI: 10.1155/2020/7865484
Phosphodiesterase 10A Is a Mediator of Osteogenic Differentiation and Mechanotransduction in Bone Marrow-Derived Mesenchymal Stromal Cells
Abstract
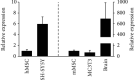
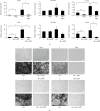
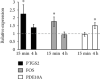
Bone marrow-derived mesenchymal stromal cells (hMSCs) are capable of differentiating into the osteogenic lineage, and for osteogenic differentiation, mechanical loading is a relevant stimulus. Mechanotransduction leads to the formation of second messengers such as cAMP, cGMP, or Ca2+ influx resulting in the activation of transcription factors mediating gene regulation. The second messengers cAMP and cGMP are degraded by phosphodiesterase isoenzymes (PDE), but the role of these enzymes during osteogenic differentiation or mechanotransduction remains unclear. Here, we focused on the isoenzyme phosphodiesterase 10A (PDE10A) and its role during osteogenic commitment and mechanotransduction. We observed a time-dependent decrease of PDE10A expression in hMSC undergoing differentiation towards the osteogenic lineage. PDE10A inhibition by papaverine diminished osteogenic differentiation. While applying mechanical strain via cyclic stretching of hMSCs led to an upregulation of PDE10A gene expression, inhibition of PDE10A using the drug papaverine repressed expression of mechanoresponsive genes. We conclude that PDE10A is a modulator of osteogenic differentiation as well as mechanotransduction in hMSCs. Our data further suggests that the relative increase of cAMP, rather than the absolute cAMP level, is a key driver of the observed effects.
Copyright © 2020 Sigrid Müller-Deubert et al.
Conflict of interest statement
There is no conflict of interest.
Figures


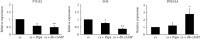
Similar articles
-
Phosphodiesterase-10A Inverse Changes in Striatopallidal and Striatoentopeduncular Pathways of a Transgenic Mouse Model of DYT1 Dystonia.J Neurosci. 2017 Feb 22;37(8):2112-2124. doi: 10.1523/JNEUROSCI.3207-15.2016. Epub 2017 Jan 23. J Neurosci. 2017. PMID: 28115486 Free PMC article.
-
Inhibition of the striatum-enriched phosphodiesterase PDE10A: a novel approach to the treatment of psychosis.Neuropharmacology. 2006 Aug;51(2):386-96. doi: 10.1016/j.neuropharm.2006.04.013. Epub 2006 Jun 15. Neuropharmacology. 2006. PMID: 16780899
-
Phosphodiesterase 10A in the rat pineal gland: localization, daily and seasonal regulation of expression and influence on signal transduction.Neuroendocrinology. 2011;94(2):113-23. doi: 10.1159/000327138. Epub 2011 Apr 7. Neuroendocrinology. 2011. PMID: 21474921
-
1-(3-[11C]Methoxy-4-methoxybenzyl)-6,7-dimethoxyisoquinoline.2010 Dec 12 [updated 2011 Mar 2]. In: Molecular Imaging and Contrast Agent Database (MICAD) [Internet]. Bethesda (MD): National Center for Biotechnology Information (US); 2004–2013. 2010 Dec 12 [updated 2011 Mar 2]. In: Molecular Imaging and Contrast Agent Database (MICAD) [Internet]. Bethesda (MD): National Center for Biotechnology Information (US); 2004–2013. PMID: 21391338 Free Books & Documents. Review.
-
Mechanisms for osteogenic differentiation of human mesenchymal stem cells induced by fluid shear stress.Biomech Model Mechanobiol. 2010 Dec;9(6):659-70. doi: 10.1007/s10237-010-0206-x. Epub 2010 Mar 23. Biomech Model Mechanobiol. 2010. PMID: 20309603 Review.
Cited by
-
Mechanoresponsive regulation of fibroblast-to-myofibroblast transition in three-dimensional tissue analogues: mechanical strain amplitude dependency of fibrosis.Sci Rep. 2022 Oct 7;12(1):16832. doi: 10.1038/s41598-022-20383-5. Sci Rep. 2022. PMID: 36207437 Free PMC article.
-
Identification of novel off targets of baricitinib and tofacitinib by machine learning with a focus on thrombosis and viral infection.Sci Rep. 2022 May 12;12(1):7843. doi: 10.1038/s41598-022-11879-1. Sci Rep. 2022. PMID: 35551258 Free PMC article.
-
Differential Effects of PTH (1-34), PTHrP (1-36), and Abaloparatide on the Murine Osteoblast Transcriptome.J Endocr Soc. 2023 Dec 13;8(1):bvad156. doi: 10.1210/jendso/bvad156. eCollection 2023 Dec 1. J Endocr Soc. 2023. PMID: 38155918 Free PMC article.
-
Growth resilience to weather variation in commercial free-ranging chickens in Ethiopia.BMC Genomics. 2025 Apr 14;26(1):371. doi: 10.1186/s12864-025-11561-6. BMC Genomics. 2025. PMID: 40229704 Free PMC article.
-
Assessment of the viability and mechanoresponsiveness of hMSC-TERT printed with bioinert, thermoresponsive hydrogels.Sci Rep. 2025 Apr 10;15(1):12257. doi: 10.1038/s41598-025-97196-9. Sci Rep. 2025. PMID: 40210996 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Miscellaneous

