Using RNA-Seq to Explore the Repair Mechanism of the Three Methods and Three-Acupoint Technique on DRGs in Sciatic Nerve Injured Rats
- PMID: 32587647
- PMCID: PMC7298348
- DOI: 10.1155/2020/7531409
Using RNA-Seq to Explore the Repair Mechanism of the Three Methods and Three-Acupoint Technique on DRGs in Sciatic Nerve Injured Rats
Abstract
Objective: To study the effects of the three methods and three-acupoint technique on DRG gene expression in SNI model rats and to elucidate the molecular mechanism of the three methods and three-acupoint technique on promoting recovery in peripheral nerve injury.
Methods: 27 male SD rats were randomly divided into three groups: a Sham group, the SNI group, and the Tuina group. The Tuina group was treated with a tuina manipulation simulator to simulate massage on points, controlling for both quality and quantity. Point-pressing, plucking, and kneading methods were administered quantitatively at Yinmen (BL37), Chengshan (BL57), and Yanglingquan (GB34) points on the affected side once a day, beginning 7 days after modeling. Intervention was applied once a day for 10 days, then 1 day of rest, followed by 10 more days of intervention, totally equaling 20 times of intervention. The effect of the three methods and three-point technique on the recovery of injured rats was evaluated using behavior analysis. RNA sequencing (RNA-Seq) analysis of differentially expressed genes in DRGs of the three groups of rats was also performed. GO and KEGG enrichment was analyzed and verified using real-time PCR.
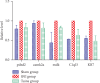
Results: RNA-Seq combined with database information showed that the number of differentially expressed genes in DRG was the largest in the Tuina group compared with the SNI group, totaling 226. GO function is enriched in the positive regulation of cell processes, ion binding, protein binding, neuron, response to pressure, response to metal ions, neuron projection, and other biological processes. GO function is also enriched in the Wnt, IL-17, and MAPK signaling pathways in the KEGG database. PCR results were consistent with those of RNA sequencing, suggesting that the results of transcriptome sequencing were reliable.
Conclusion: The three methods and three-acupoint technique can promote the recovery of SNI model rats by altering the gene sequence in DRGs.
Copyright © 2020 Tao-tao Lv et al.
Conflict of interest statement
The authors declare that there are no conflicts of interest.
Figures



References
-
- Tölle T. R. Challenges with current treatment of neuropathicpain. European Journal of Pain Supplements. 2010;4(2):161–165. doi: 10.1016/s1754-3207(10)70527-7. - DOI
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

