A Novel, Rapid, and Simple PMA-qPCR Method for Detection and Counting of Viable Brucella Organisms
- PMID: 32587912
- PMCID: PMC7305652
- DOI: 10.2478/jvetres-2020-0033
A Novel, Rapid, and Simple PMA-qPCR Method for Detection and Counting of Viable Brucella Organisms
Abstract
Introduction: The plate counting method widely used at present to discern viable from non-viable Brucella in the host or cell is time-consuming and laborious. Therefore, it is necessary to establish a rapid, simple method for detecting and counting viable Brucella organisms.
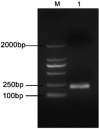
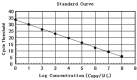
Material and methods: Using propidium monoazide (PMA) to inhibit amplification of DNA from dead Brucella, a novel, rapid PMA-quantitative PCR (PMA-qPCR) detection method for counting viable Brucella was established. The standard recombinant plasmid with the target BCSP31 gene fragment inserted was constructed for drawing a standard curve. The reaction conditions were optimised, and the sensitivity, specificity, and repeatability were analysed.
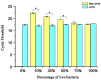
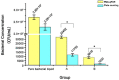
Results: The optimal exposure time and working concentration of PMA were 10 min and 15 μg/mL, respectively. The correlation coefficient (R2) of the standard curve was 0.999. The sensitivity of the method was 103 CFU/mL, moreover, its specificity and repeatability also met the requirements. The concentration of B. suis measured by the PMA-qPCR did not differ significantly from that measured by the plate counting method, and the concentrations of viable bacteria in infected cells determined by the two methods were of the same order of magnitude.
Conclusion: In this study, a rapid and simple PMA-qPCR counting method for viable Brucella was established, which will facilitate related research.
Keywords: BCSP31gene; Brucella; propidium monoazide; quantitative PCR; viable bacteria.
© 2020 S.J. Zhang et al. published by Sciendo.
Conflict of interest statement
Conflict of Interest Conflict of Interests Statement: The authors declare that there is no conflict of interests regarding the publication of this article.
Figures



References
-
- Arenas-Gamboa A.M., Ficht T.A., Kahl-McDonagh M.M., Gomez G., Rice-Ficht A.C.. The Brucella abortus S19 DeltavjbR live vaccine candidate is safer than S19 and confers protection against wild-type challenge in BALB/c mice when delivered in a sustained-release vehicle. Infect Immun. 2009;77:877–884. - PMC - PubMed
-
- Baily G.G., Krahn J.B., Drasar B.S., Stoker N.G.. Detection of Brucella melitensis and Brucella abortus by DNA amplification. J Trop Med Hyg. 1992;95:271–275. - PubMed
-
- Bounaadja L., Albert D., Chenais B., Henault S., Zygmunt M.S., Poliak S., Garin-Bastuji B.. Real-time PCR for identification of Brucella spp.: a comparative study of IS711, bcsp31 and per target genes. Vet Microbiol. 2009;137:156–164. - PubMed
-
- Chang C.W., Lin M.H.. Optimization of PMA-qPCR for Staphylococcus aureus and determination of viable bacteria in indoor air. Indoor Air. 2018;28:64–72. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
