Understanding the Interaction of Polyelectrolyte Architectures with Proteins and Biosystems
- PMID: 32589355
- PMCID: PMC7894192
- DOI: 10.1002/anie.202006457
Understanding the Interaction of Polyelectrolyte Architectures with Proteins and Biosystems
Abstract
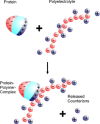
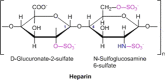
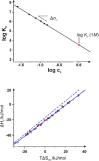
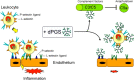
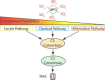
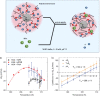
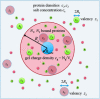
The counterions neutralizing the charges on polyelectrolytes such as DNA or heparin may dissociate in water and greatly influence the interaction of such polyelectrolytes with biomolecules, particularly proteins. In this Review we give an overview of studies on the interaction of proteins with polyelectrolytes and how this knowledge can be used for medical applications. Counterion release was identified as the main driving force for the binding of proteins to polyelectrolytes: Patches of positive charge become multivalent counterions of the polyelectrolyte and lead to the release of counterions from the polyelectrolyte and a concomitant increase in entropy. This is shown from investigations on the interaction of proteins with natural and synthetic polyelectrolytes. Special emphasis is paid to sulfated dendritic polyglycerols (dPGS). The Review demonstrates that we are moving to a better understanding of charge-charge interactions in systems of biological relevance. Research along these lines will aid and promote the design of synthetic polyelectrolytes for medical applications.
Keywords: complementary binding; counterion release; heparin; inflammation; polyelectrolytes.
© 2020 The Authors. Published by Wiley-VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

