Comparative genomics of the genus Roseburia reveals divergent biosynthetic pathways that may influence colonic competition among species
- PMID: 32589566
- PMCID: PMC7478625
- DOI: 10.1099/mgen.0.000399
Comparative genomics of the genus Roseburia reveals divergent biosynthetic pathways that may influence colonic competition among species
Abstract
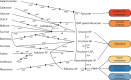
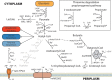
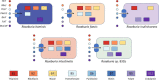
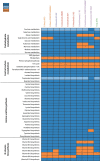
Roseburia species are important denizens of the human gut microbiome that ferment complex polysaccharides to butyrate as a terminal fermentation product, which influences human physiology and serves as an energy source for colonocytes. Previous comparative genomics analyses of the genus Roseburia have examined polysaccharide degradation genes. Here, we characterize the core and pangenomes of the genus Roseburia with respect to central carbon and energy metabolism, as well as biosynthesis of amino acids and B vitamins using orthology-based methods, uncovering significant differences among species in their biosynthetic capacities. Variation in gene content among Roseburia species and strains was most significant for cofactor biosynthesis. Unlike all other species of Roseburia that we analysed, Roseburia inulinivorans strains lacked biosynthetic genes for riboflavin or pantothenate but possessed folate biosynthesis genes. Differences in gene content for B vitamin synthesis were matched with differences in putative salvage and synthesis strategies among species. For example, we observed extended biotin salvage capabilities in R. intestinalis strains, which further suggest that B vitamin acquisition strategies may impact fitness in the gut ecosystem. As differences in the functional potential to synthesize components of biomass (e.g. amino acids, vitamins) can drive interspecies interactions, variation in auxotrophies of the Roseburia spp. genomes may influence in vivo gut ecology. This study serves to advance our understanding of the potential metabolic interactions that influence the ecology of Roseburia spp. and, ultimately, may provide a basis for rational strategies to manipulate the abundances of these species.
Keywords: B vitamin biosynthesis; Lachnospiraceae; Roseburia; amino acid biosynthesis; butyrate synthesis; comparative genomics.
Conflict of interest statement
The authors declare that there are no conflicts of interest.
Figures


References
-
- Liu C, Finegold SM, Song Y, Lawson PA. Reclassification of Clostridium coccoides, Ruminococcus hansenii, Ruminococcus hydrogenotrophicus, Ruminococcus luti, Ruminococcus productus and Ruminococcus schinkii as Blautia coccoides gen. nov., comb. nov. Blautia hansenii comb. nov., Blaut. Int J Syst Evol Microbiol. 2008;58:1896–1902. - PubMed
-
- Tamanai-Shacoori Z, Smida I, Bousarghin L, Loreal O, Meuric V, et al. Roseburia spp.: a marker of health? Future Microbiol. 2017;12:157–170. - PubMed
-
- Rosero JA, Killer J, Sechovcová H, Mrázek J, Benada O, et al. Reclassification of Eubacterium rectale (Hauduroy, et al. 1937) Prévot 1938 in a new genus Agathobacter gen. nov. as Agathobacter rectalis comb. nov., and description of Agathobacter ruminis sp. nov., isolated from the rumen contents of sheep and cows. Int J Syst Evol Microbiol. 2016;66:768–773. - PubMed
-
- Scott KP, Duncan SH, Flint HJ. Dietary fibre and the gut microbiota. Nutr Bull. 2008;33:201–211.
-
- Duncan SH, Aminov RI, Scott KP, Louis P, Stanton TB, et al. Proposal of Roseburia faecis sp. nov., Roseburia hominis sp. nov. and Roseburia inulinivorans sp. nov., based on isolates from human faeces. Int J Syst Evol Microbiol. 2006:2437–2441. - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

