c-di-GMP inhibits LonA-dependent proteolysis of TfoY in Vibrio cholerae
- PMID: 32589664
- PMCID: PMC7371385
- DOI: 10.1371/journal.pgen.1008897
c-di-GMP inhibits LonA-dependent proteolysis of TfoY in Vibrio cholerae
Abstract
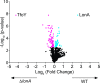
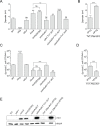
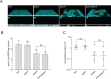
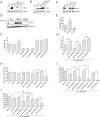
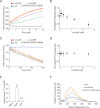
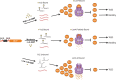
The LonA (or Lon) protease is a central post-translational regulator in diverse bacterial species. In Vibrio cholerae, LonA regulates a broad range of behaviors including cell division, biofilm formation, flagellar motility, c-di-GMP levels, the type VI secretion system (T6SS), virulence gene expression, and host colonization. Despite LonA's role in cellular processes critical for V. cholerae's aquatic and infectious life cycles, relatively few LonA substrates have been identified. LonA protease substrates were therefore identified through comparison of the proteomes of wild-type and ΔlonA strains following translational inhibition. The most significantly enriched LonA-dependent protein was TfoY, a known regulator of motility and the T6SS in V. cholerae. Experiments showed that TfoY was required for LonA-mediated repression of motility and T6SS-dependent killing. In addition, TfoY was stabilized under high c-di-GMP conditions and biochemical analysis determined direct binding of c-di-GMP to LonA results in inhibition of its protease activity. The work presented here adds to the list of LonA substrates, identifies LonA as a c-di-GMP receptor, demonstrates that c-di-GMP regulates LonA activity and TfoY protein stability, and helps elucidate the mechanisms by which LonA controls important V. cholerae behaviors.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

Similar articles
-
The LonA Protease Regulates Biofilm Formation, Motility, Virulence, and the Type VI Secretion System in Vibrio cholerae.J Bacteriol. 2016 Jan 11;198(6):973-85. doi: 10.1128/JB.00741-15. J Bacteriol. 2016. PMID: 26755629 Free PMC article.
-
Cyclic di-GMP Regulates TfoY in Vibrio cholerae To Control Motility by both Transcriptional and Posttranscriptional Mechanisms.J Bacteriol. 2018 Mar 12;200(7):e00578-17. doi: 10.1128/JB.00578-17. Print 2018 Apr 1. J Bacteriol. 2018. PMID: 29311281 Free PMC article.
-
A systematic analysis of the in vitro and in vivo functions of the HD-GYP domain proteins of Vibrio cholerae.BMC Microbiol. 2014 Oct 25;14:272. doi: 10.1186/s12866-014-0272-9. BMC Microbiol. 2014. PMID: 25343965 Free PMC article.
-
The ins and outs of cyclic di-GMP signaling in Vibrio cholerae.Curr Opin Microbiol. 2017 Apr;36:20-29. doi: 10.1016/j.mib.2017.01.002. Epub 2017 Feb 5. Curr Opin Microbiol. 2017. PMID: 28171809 Free PMC article. Review.
-
Rules of Engagement: The Type VI Secretion System in Vibrio cholerae.Trends Microbiol. 2017 Apr;25(4):267-279. doi: 10.1016/j.tim.2016.12.003. Epub 2016 Dec 24. Trends Microbiol. 2017. PMID: 28027803 Free PMC article. Review.
Cited by
-
Structural and mechanistic studies on human LONP1 redefine the hand-over-hand translocation mechanism.bioRxiv [Preprint]. 2024 Jun 25:2024.06.24.600538. doi: 10.1101/2024.06.24.600538. bioRxiv. 2024. PMID: 38979310 Free PMC article. Preprint.
-
Regulation of type VI secretion systems at the transcriptional, posttranscriptional and posttranslational level.Microbiology (Reading). 2023 Aug;169(8):001376. doi: 10.1099/mic.0.001376. Microbiology (Reading). 2023. PMID: 37552221 Free PMC article. Review.
-
Cyclic Diguanylate in the Wild: Roles During Plant and Animal Colonization.Annu Rev Microbiol. 2024 Nov;78(1):533-551. doi: 10.1146/annurev-micro-041522-101729. Epub 2024 Nov 7. Annu Rev Microbiol. 2024. PMID: 39270684 Free PMC article. Review.
-
Type VI Secretion Systems: Environmental and Intra-host Competition of Vibrio cholerae.Adv Exp Med Biol. 2023;1404:41-63. doi: 10.1007/978-3-031-22997-8_3. Adv Exp Med Biol. 2023. PMID: 36792870
-
Evolution of cyclic di-GMP signalling on a short and long term time scale.Microbiology (Reading). 2023 Jun;169(6):001354. doi: 10.1099/mic.0.001354. Microbiology (Reading). 2023. PMID: 37384391 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

