System-wide biochemical analysis reveals ozonide antimalarials initially act by disrupting Plasmodium falciparum haemoglobin digestion
- PMID: 32589689
- PMCID: PMC7347234
- DOI: 10.1371/journal.ppat.1008485
System-wide biochemical analysis reveals ozonide antimalarials initially act by disrupting Plasmodium falciparum haemoglobin digestion
Abstract
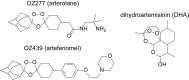
Ozonide antimalarials, OZ277 (arterolane) and OZ439 (artefenomel), are synthetic peroxide-based antimalarials with potent activity against the deadliest malaria parasite, Plasmodium falciparum. Here we used a "multi-omics" workflow, in combination with activity-based protein profiling (ABPP), to demonstrate that peroxide antimalarials initially target the haemoglobin (Hb) digestion pathway to kill malaria parasites. Time-dependent metabolomic profiling of ozonide-treated P. falciparum infected red blood cells revealed a rapid depletion of short Hb-derived peptides followed by subsequent alterations in lipid and nucleotide metabolism, while untargeted peptidomics showed accumulation of longer Hb-derived peptides. Quantitative proteomics and ABPP assays demonstrated that Hb-digesting proteases were increased in abundance and activity following treatment, respectively. Ozonide-induced depletion of short Hb-derived peptides was less extensive in a drug-treated K13-mutant artemisinin resistant parasite line (Cam3.IIR539T) than in the drug-treated isogenic sensitive strain (Cam3.IIrev), further confirming the association between ozonide activity and Hb catabolism. To demonstrate that compromised Hb catabolism may be a primary mechanism involved in ozonide antimalarial activity, we showed that parasites forced to rely solely on Hb digestion for amino acids became hypersensitive to short ozonide exposures. Quantitative proteomics analysis also revealed parasite proteins involved in translation and the ubiquitin-proteasome system were enriched following drug treatment, suggestive of the parasite engaging a stress response to mitigate ozonide-induced damage. Taken together, these data point to a mechanism of action involving initial impairment of Hb catabolism, and indicate that the parasite regulates protein turnover to manage ozonide-induced damage.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


Similar articles
-
Parasite-Mediated Degradation of Synthetic Ozonide Antimalarials Impacts In Vitro Antimalarial Activity.Antimicrob Agents Chemother. 2018 Feb 23;62(3):e01566-17. doi: 10.1128/AAC.01566-17. Print 2018 Mar. Antimicrob Agents Chemother. 2018. PMID: 29263074 Free PMC article.
-
Plasmodium falciparum K13 Mutations Differentially Impact Ozonide Susceptibility and Parasite Fitness In Vitro.mBio. 2017 Apr 11;8(2):e00172-17. doi: 10.1128/mBio.00172-17. mBio. 2017. PMID: 28400526 Free PMC article.
-
In vitro activity of anti-malarial ozonides against an artemisinin-resistant isolate.Malar J. 2017 Jan 25;16(1):45. doi: 10.1186/s12936-017-1696-0. Malar J. 2017. PMID: 28122617 Free PMC article.
-
Ozonide Antimalarial Activity in the Context of Artemisinin-Resistant Malaria.Trends Parasitol. 2019 Jul;35(7):529-543. doi: 10.1016/j.pt.2019.05.002. Epub 2019 Jun 5. Trends Parasitol. 2019. PMID: 31176584 Review.
-
Considerations on the mechanism of action of artemisinin antimalarials: part 1--the 'carbon radical' and 'heme' hypotheses.Infect Disord Drug Targets. 2013 Aug;13(4):217-77. doi: 10.2174/1871526513666131129155708. Infect Disord Drug Targets. 2013. PMID: 24304352 Review.
Cited by
-
Genetic and chemical validation of Plasmodium falciparum aminopeptidase PfA-M17 as a drug target in the hemoglobin digestion pathway.Elife. 2022 Sep 13;11:e80813. doi: 10.7554/eLife.80813. Elife. 2022. PMID: 36097817 Free PMC article.
-
Natural Products That Changed Society.Biomedicines. 2021 Apr 26;9(5):472. doi: 10.3390/biomedicines9050472. Biomedicines. 2021. PMID: 33925870 Free PMC article. Review.
-
Genetic complexity alters drug susceptibility of asexual and gametocyte stages of Plasmodium falciparum to antimalarial candidates.Antimicrob Agents Chemother. 2024 Mar 6;68(3):e0129123. doi: 10.1128/aac.01291-23. Epub 2024 Jan 23. Antimicrob Agents Chemother. 2024. PMID: 38259087 Free PMC article.
-
Adapt or Die: Targeting Unique Transmission-Stage Biology for Malaria Elimination.Front Cell Infect Microbiol. 2022 Jun 9;12:901971. doi: 10.3389/fcimb.2022.901971. eCollection 2022. Front Cell Infect Microbiol. 2022. PMID: 35755845 Free PMC article. Review.
-
Chemoproteomics validates selective targeting of Plasmodium M1 alanyl aminopeptidase as an antimalarial strategy.Elife. 2024 Jul 8;13:RP92990. doi: 10.7554/eLife.92990. Elife. 2024. PMID: 38976500 Free PMC article.
References
-
- WHO. World Malaria Report 2017. Geneva: World Health Organisation, 2017.
-
- Posner GH, Oh CH. Regiospecifically oxygen-18 labeled 1, 2, 4-trioxane: a simple chemical model system to probe the mechanism (s) for the antimalarial activity of artemisinin (qinghaosu). J Am Chem Soc. 1992; 114: 8328–9. 10.1021/ja00047a076 - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

