Selective Translation Complex Profiling Reveals Staged Initiation and Co-translational Assembly of Initiation Factor Complexes
- PMID: 32589964
- PMCID: PMC7447980
- DOI: 10.1016/j.molcel.2020.06.004
Selective Translation Complex Profiling Reveals Staged Initiation and Co-translational Assembly of Initiation Factor Complexes
Abstract
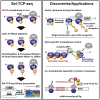
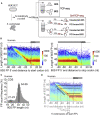
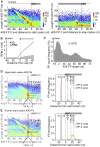
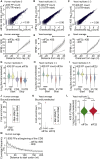
Translational control targeting the initiation phase is central to the regulation of gene expression. Understanding all of its aspects requires substantial technological advancements. Here we modified yeast translation complex profile sequencing (TCP-seq), related to ribosome profiling, and adapted it for mammalian cells. Human TCP-seq, capable of capturing footprints of 40S subunits (40Ss) in addition to 80S ribosomes (80Ss), revealed that mammalian and yeast 40Ss distribute similarly across 5'TRs, indicating considerable evolutionary conservation. We further developed yeast and human selective TCP-seq (Sel-TCP-seq), enabling selection of 40Ss and 80Ss associated with immuno-targeted factors. Sel-TCP-seq demonstrated that eIF2 and eIF3 travel along 5' UTRs with scanning 40Ss to successively dissociate upon AUG recognition; notably, a proportion of eIF3 lingers on during the initial elongation cycles. Highlighting Sel-TCP-seq versatility, we also identified four initiating 48S conformational intermediates, provided novel insights into ATF4 and GCN4 mRNA translational control, and demonstrated co-translational assembly of initiation factor complexes.
Keywords: ATF4; GCN4; Ribo-seq; TCP-seq; UTR; co-translational assembly; eIF2; eIF3; gene expression; mRNA; ribosome; ribosome profiling; translational control.
Copyright © 2020 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Interests The authors declare no competing interests.
Figures



Comment in
-
Should I Stay or Should I Go: eIF3 Remains Ribosome Associated and Is Required for Elongation.Mol Cell. 2020 Aug 20;79(4):539-541. doi: 10.1016/j.molcel.2020.07.025. Mol Cell. 2020. PMID: 32822578
References
-
- Archer S.K., Shirokikh N.E., Beilharz T.H., Preiss T. Dynamics of ribosome scanning and recycling revealed by translation complex profiling. Nature. 2016;535:570–574. - PubMed
-
- Asano K., Phan L., Anderson J., Hinnebusch A.G. Complex formation by all five homologues of mammalian translation initiation factor 3 subunits from yeast Saccharomyces cerevisiae. J. Biol. Chem. 1998;273:18573–18585. - PubMed
-
- Asano K., Phan L., Valásek L., Schoenfeld L.W., Shalev A., Clayton J., Nielsen K., Donahue T.F., Hinnebusch A.G. A multifactor complex of eIF1, eIF2, eIF3, eIF5, and tRNA(i)Met promotes initiation complex assembly and couples GTP hydrolysis to AUG recognition. Cold Spring Harb. Symp. Quant. Biol. 2001;66:403–415. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

