Developing standards for the microbiome field
- PMID: 32591016
- PMCID: PMC7320585
- DOI: 10.1186/s40168-020-00856-3
Developing standards for the microbiome field
Abstract
Background: Effective standardisation of methodologies to analyse the microbiome is essential to the entire microbiome community. Despite the microbiome field being established for over a decade, there are no accredited or certified reference materials available to the wider community. In this study, we describe the development of the first reference reagents produced by the National Institute for Biological Standards and Control (NIBSC) for microbiome analysis by next-generation sequencing. These can act as global working standards and will be evaluated as candidate World Health Organization International Reference Reagents.
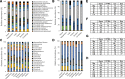
Results: We developed the NIBSC DNA reference reagents Gut-Mix-RR and Gut-HiLo-RR and a four-measure framework for evaluation of bioinformatics tool and pipeline bias. Using these reagents and reporting system, we performed an independent evaluation of a variety of bioinformatics tools by analysing shotgun sequencing and 16S rRNA sequencing data generated from the Gut-Mix-RR and Gut-HiLo-RR. We demonstrate that key measures of microbiome health, such as diversity estimates, are largely inflated by the majority of bioinformatics tools. Across all tested tools, biases were present, with a clear trade-off occurring between sensitivity and the relative abundance of false positives in the final dataset. Using commercially available mock communities, we investigated how the composition of reference reagents may impact benchmarking studies. Reporting measures consistently changed when the same bioinformatics tools were used on different community compositions. This was influenced by both community complexity and taxonomy of species present. Both NIBSC reference reagents, which consisted of gut commensal species, proved to be the most challenging for the majority of bioinformatics tools tested. Going forward, we recommend the field uses site-specific reagents of a high complexity to ensure pipeline benchmarking is fit for purpose.
Conclusions: If a consensus of acceptable levels of error can be agreed on, widespread adoption of these reference reagents will standardise downstream gut microbiome analyses. We propose to do this through a large open-invite collaborative study for multiple laboratories in 2020. Video Abstract.
Conflict of interest statement
NIBSC is a centre of the UK’s Medicine and Healthcare product Regulatory Agency and is a non-profit institute which generates global reference reagents for public health. Reagents generated by NIBSC will be published here will be distributed on a cost-recovery basis. We have no other competing interests to declare.
Figures


References
-
- Sinha R, Abu-Ali G, Vogtmann E, Fodor AA, Ren B, Amir A, Schwager E, Crabtree J, Ma S. Microbiome Quality Control Project C et al: Assessment of variation in microbial community amplicon sequencing by the Microbiome Quality Control (MBQC) project consortium. Nat Biotechnol. 2017;35(11):1077–1086. doi: 10.1038/nbt.3981. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

