Maternal factors regulating preimplantation development in mice
- PMID: 32591079
- PMCID: PMC8394711
- DOI: 10.1016/bs.ctdb.2019.10.006
Maternal factors regulating preimplantation development in mice
Abstract
Mammalian embryogenesis depends on maternal factors accumulated in eggs prior to fertilization and on placental transfers later in gestation. In this review, we focus on initial events when the organism has insufficient newly synthesized embryonic factors to sustain development. These maternal factors regulate preimplantation embryogenesis both uniquely in pronuclear formation, genome reprogramming and cell fate determination and more universally in regulating cell division, transcription and RNA metabolism. Depletion, disruption or inappropriate persistence of maternal factors can result in developmental defects in early embryos. To better understand the origins of these maternal effects, we include oocyte maturation processes that are responsible for their production. We focus on recent publications and reference comprehensive reviews that include earlier scientific literature of early mouse development.
Keywords: Maternal RNA degradation; Preimplantation embryonic development; Zygotic genome activation.
© 2020 Elsevier Inc. All rights reserved.
Figures
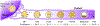
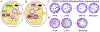
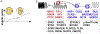
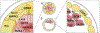

References
-
- Aoki F, Worrad DM, & Schultz RM (1997). Regulation of transcriptional activity during the first and second cell cycles in the preimplantation mouse embryo. Developmental Biology, 181, 296–307. - PubMed
-
- Baran V, Solc P, Kovarikova V, Rehak P, & Sutovsky P (2013). Polo-like kinase 1 is essential for the first mitotic division in the mouse embryo. Molecular Reproduction and Development, 80, 522–534. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

