Genetic drug target validation using Mendelian randomisation
- PMID: 32591531
- PMCID: PMC7320010
- DOI: 10.1038/s41467-020-16969-0
Genetic drug target validation using Mendelian randomisation
Abstract
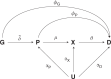
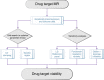
Mendelian randomisation (MR) analysis is an important tool to elucidate the causal relevance of environmental and biological risk factors for disease. However, causal inference is undermined if genetic variants used to instrument a risk factor also influence alternative disease-pathways (horizontal pleiotropy). Here we report how the 'no horizontal pleiotropy assumption' is strengthened when proteins are the risk factors of interest. Proteins are typically the proximal effectors of biological processes encoded in the genome. Moreover, proteins are the targets of most medicines, so MR studies of drug targets are becoming a fundamental tool in drug development. To enable such studies, we introduce a mathematical framework that contrasts MR analysis of proteins with that of risk factors located more distally in the causal chain from gene to disease. We illustrate key model decisions and introduce an analytical framework for maximising power and evaluating the robustness of analyses.
Conflict of interest statement
DFF is a full-time employee of Bayer AG, Germany. BT is a full-time employee of Servier. RSP has received honoraria from Sanofi, Bayer and Amgen. MZ is a full-time employee of GSK. AFS and FWA have received Servier funding for unrelated work. MZ conducted this research as an employee of BenevolentAI. Since completing the work MZ is now a full-time employee of GlaxoSmithKline. None of the remaining authors have a competing interest to declare.
Figures



References
-
- Hingorani A, Humphries S. Nature’s randomised trials. Lancet. 2005;366:1906–1908. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

