MRI signatures of brain age and disease over the lifespan based on a deep brain network and 14 468 individuals worldwide
- PMID: 32591831
- PMCID: PMC7364766
- DOI: 10.1093/brain/awaa160
MRI signatures of brain age and disease over the lifespan based on a deep brain network and 14 468 individuals worldwide
Erratum in
-
Corrigendum to: MRI signatures of brain age and disease over the lifespan based on a deep brain network and 14 468 individuals worldwide.Brain. 2021 Feb 12;144(1):e12. doi: 10.1093/brain/awaa328. Brain. 2021. PMID: 33057608 Free PMC article. No abstract available.
Abstract
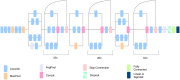
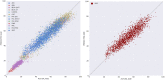
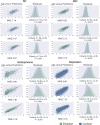
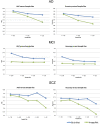
Deep learning has emerged as a powerful approach to constructing imaging signatures of normal brain ageing as well as of various neuropathological processes associated with brain diseases. In particular, MRI-derived brain age has been used as a comprehensive biomarker of brain health that can identify both advanced and resilient ageing individuals via deviations from typical brain ageing. Imaging signatures of various brain diseases, including schizophrenia and Alzheimer's disease, have also been identified using machine learning. Prior efforts to derive these indices have been hampered by the need for sophisticated and not easily reproducible processing steps, by insufficiently powered or diversified samples from which typical brain ageing trajectories were derived, and by limited reproducibility across populations and MRI scanners. Herein, we develop and test a sophisticated deep brain network (DeepBrainNet) using a large (n = 11 729) set of MRI scans from a highly diversified cohort spanning different studies, scanners, ages and geographic locations around the world. Tests using both cross-validation and a separate replication cohort of 2739 individuals indicate that DeepBrainNet obtains robust brain-age estimates from these diverse datasets without the need for specialized image data preparation and processing. Furthermore, we show evidence that moderately fit brain ageing models may provide brain age estimates that are most discriminant of individuals with pathologies. This is not unexpected as tightly-fitting brain age models naturally produce brain-age estimates that offer little information beyond age, and loosely fitting models may contain a lot of noise. Our results offer some experimental evidence against commonly pursued tightly-fitting models. We show that the moderately fitting brain age models obtain significantly higher differentiation compared to tightly-fitting models in two of the four disease groups tested. Critically, we demonstrate that leveraging DeepBrainNet, along with transfer learning, allows us to construct more accurate classifiers of several brain diseases, compared to directly training classifiers on patient versus healthy control datasets or using common imaging databases such as ImageNet. We, therefore, derive a domain-specific deep network likely to reduce the need for application-specific adaptation and tuning of generic deep learning networks. We made the DeepBrainNet model freely available to the community for MRI-based evaluation of brain health in the general population and over the lifespan.
Keywords: brain age; deep learning; transfer learning.
© The Author(s) (2020). Published by Oxford University Press on behalf of the Guarantors of Brain. All rights reserved. For permissions, please email: journals.permissions@oup.com.
Figures
Comment in
-
Beyond the AJR: "MRI Signatures of Brain Age and Disease Over the Lifespan Based on a Deep Brain Network and 14 468 Individuals Worldwide".AJR Am J Roentgenol. 2021 May;216(5):1170. doi: 10.2214/AJR.20.24985. Epub 2020 Oct 28. AJR Am J Roentgenol. 2021. PMID: 33112202 No abstract available.
-
Reply: From 'loose fitting' to high-performance, uncertainty-aware brain-age modelling.Brain. 2021 Apr 12;144(3):e32. doi: 10.1093/brain/awaa455. Brain. 2021. PMID: 33826693 No abstract available.
-
From 'loose fitting' to high-performance, uncertainty-aware brain-age modelling.Brain. 2021 Apr 12;144(3):e31. doi: 10.1093/brain/awaa454. Brain. 2021. PMID: 33826702 No abstract available.
References
-
- Abadi M, Barham P, Chen J, Chen Z, Davis A, Dean J, et al. Tensorflow: A system for large-scale machine learning. In: 12th USENIX Symposium on Operating Systems Design and Implementation (OSDI 16), 2016. P. 265–83.
-
- Anwar SM, Majid M, Qayyum A, Awais M, Alnowami M, Khan MK.. Medical image analysis using convolutional neural networks: a review. J Med Syst 2018; 42: 226. - PubMed
-
- Becherer N. Transfer Learning in Convolutional Neural Networks for Fine-Grained Image Classification; 2017.
Publication types
MeSH terms
Grants and funding
- R01 EB022573/EB/NIBIB NIH HHS/United States
- R01 AG027161/AG/NIA NIH HHS/United States
- R01 AG021155/AG/NIA NIH HHS/United States
- RF1 AG027161/AG/NIA NIH HHS/United States
- RF1 AG059869/AG/NIA NIH HHS/United States
- RF1 AG054409/AG/NIA NIH HHS/United States
- S10 OD023495/OD/NIH HHS/United States
- P30 AG062715/AG/NIA NIH HHS/United States
- R01 HL127659/HL/NHLBI NIH HHS/United States
- P30 AG066507/AG/NIA NIH HHS/United States
- HHSN271201600059C/DA/NIDA NIH HHS/United States
- R01 MH112070/MH/NIMH NIH HHS/United States
- U19 AG033655/AG/NIA NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

