An Insulin-Sensitive Circular RNA that Regulates Lifespan in Drosophila
- PMID: 32592682
- PMCID: PMC7318944
- DOI: 10.1016/j.molcel.2020.06.011
An Insulin-Sensitive Circular RNA that Regulates Lifespan in Drosophila
Abstract
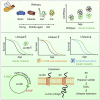
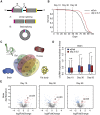
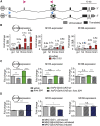
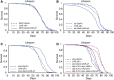
Circular RNAs (circRNAs) are abundant and accumulate with age in neurons of diverse species. However, only few circRNAs have been functionally characterized, and their role during aging has not been addressed. Here, we use transcriptome profiling during aging and find that accumulation of circRNAs is slowed down in long-lived insulin mutant flies. Next, we characterize the in vivo function of a circRNA generated by the sulfateless gene (circSfl), which is consistently upregulated, particularly in the brain and muscle, of diverse long-lived insulin mutants. Strikingly, lifespan extension of insulin mutants is dependent on circSfl, and overexpression of circSfl alone is sufficient to extend the lifespan. Moreover, circSfl is translated into a protein that shares the N terminus and potentially some functions with the full-length Sfl protein encoded by the host gene. Our study demonstrates that insulin signaling affects global circRNA accumulation and reveals an important role of circSfl during aging in vivo.
Keywords: Drosophila; ageing; alternative splicing; backsplicing; circRNA; heparan sulfate; insulin; longevity; non-coding RNAs; sulfateless.
Copyright © 2020 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Interests The authors declare no competing interests.
Figures




Comment in
-
CircRNAs in lifespan.Nat Rev Mol Cell Biol. 2020 Aug;21(8):420. doi: 10.1038/s41580-020-0269-1. Nat Rev Mol Cell Biol. 2020. PMID: 32616909 No abstract available.
References
-
- Ashwal-Fluss R., Meyer M., Pamudurti N.R., Ivanov A., Bartok O., Hanan M., Evantal N., Memczak S., Rajewsky N., Kadener S. circRNA biogenesis competes with pre-mRNA splicing. Mol. Cell. 2014;56:55–66. - PubMed
-
- Baeg G.H., Selva E.M., Goodman R.M., Dasgupta R., Perrimon N. The Wingless morphogen gradient is established by the cooperative action of Frizzled and Heparan Sulfate Proteoglycan receptors. Dev. Biol. 2004;276:89–100. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

