Microbiomes of Caribbean Octocorals Vary Over Time but Are Resistant to Environmental Change
- PMID: 32595627
- PMCID: PMC7304229
- DOI: 10.3389/fmicb.2020.01272
Microbiomes of Caribbean Octocorals Vary Over Time but Are Resistant to Environmental Change
Abstract
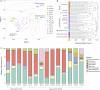
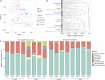
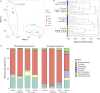
The bacterial microbiome is an essential component of many corals, although knowledge of the microbiomes in scleractinian corals far exceeds that for octocorals. This study characterized the bacterial communities present in shallow water Caribbean gorgonian octocorals over time and space, in addition to determining the bacterial assemblages in gorgonians exposed to environmental perturbations. We found that seven shallow water Caribbean gorgonian species maintained distinct microbiomes and predominantly harbored two bacterial genera, Mycoplasma and Endozoicomonas. Representatives of these taxa accounted for over 70% of the sequences recovered, made up the three most common operational taxonomic units (OTUs), and were present in most of the gorgonian species. Gorgonian species sampled in different seasons and/or in different years, exhibited significant shifts in the abundances of these bacterial OTUs, though there were few changes to overall bacterial diversity, or to the specific OTUs present. These shifts had minimal impact on the relative abundance of inferred functional proteins within the gorgonian corals. Sequences identified as Escherichia were ubiquitous in gorgonian colonies sampled from a lagoon but not in colonies sampled from a back reef. Exposure to increased temperature and/or ultraviolet radiation (UVR) or nutrient enrichment led to few significant changes in the gorgonian coral microbiomes. While there were some shifts in the abundance of the prevalent bacteria, more commonly observed was "microbial switching" between different OTUs identified within the same bacterial genus. The relative stability of gorgonian coral bacterial microbiome may potentially explain some of the resistance and resilience of Caribbean gorgonian corals against changing environmental conditions.
Keywords: UVR; bacteria; coral; gorgonian; microbiome; nitrogen; phosphorus; temperature.
Copyright © 2020 McCauley, Jackson and Goulet.
Figures



Similar articles
-
Microbiomes of stony and soft deep-sea corals share rare core bacteria.Microbiome. 2019 Jun 10;7(1):90. doi: 10.1186/s40168-019-0697-3. Microbiome. 2019. PMID: 31182168 Free PMC article.
-
Symbiodinium photosynthesis in Caribbean octocorals.PLoS One. 2014 Sep 5;9(9):e106419. doi: 10.1371/journal.pone.0106419. eCollection 2014. PLoS One. 2014. PMID: 25192405 Free PMC article.
-
Comparative Assessment of Mediterranean Gorgonian-Associated Microbial Communities Reveals Conserved Core and Locally Variant Bacteria.Microb Ecol. 2017 Feb;73(2):466-478. doi: 10.1007/s00248-016-0858-x. Epub 2016 Oct 10. Microb Ecol. 2017. PMID: 27726033
-
Bacterial associates of two Caribbean coral species reveal species-specific distribution and geographic variability.Appl Environ Microbiol. 2012 Sep;78(18):6438-49. doi: 10.1128/AEM.01162-12. Epub 2012 Jul 6. Appl Environ Microbiol. 2012. PMID: 22773636 Free PMC article.
-
The natural products chemistry of West Indian gorgonian octocorals.Tetrahedron. 1995 Apr 17;51(16):4571-4618. doi: 10.1016/0040-4020(95)00216-U. Epub 2001 Feb 1. Tetrahedron. 1995. PMID: 32287414 Free PMC article. Review. No abstract available.
Cited by
-
Towards enhancing coral heat tolerance: a "microbiome transplantation" treatment using inoculations of homogenized coral tissues.Microbiome. 2021 May 6;9(1):102. doi: 10.1186/s40168-021-01053-6. Microbiome. 2021. PMID: 33957989 Free PMC article.
-
Responses of Symbiodiniaceae Shuffling and Microbial Community Assembly in Thermally Stressed Acropora hyacinthus.Front Microbiol. 2022 Apr 1;13:832081. doi: 10.3389/fmicb.2022.832081. eCollection 2022. Front Microbiol. 2022. PMID: 35432258 Free PMC article.
-
Systematic review of cnidarian microbiomes reveals insights into the structure, specificity, and fidelity of marine associations.Nat Commun. 2023 Aug 14;14(1):4899. doi: 10.1038/s41467-023-39876-6. Nat Commun. 2023. PMID: 37580316 Free PMC article.
-
Coral microbiomes are structured by environmental gradients in deep waters.Environ Microbiome. 2024 Jun 10;19(1):38. doi: 10.1186/s40793-024-00579-0. Environ Microbiome. 2024. PMID: 38858739 Free PMC article.
-
Microbial shifts associated to ENSO-derived thermal anomalies reveal coral acclimation at holobiont level.Sci Rep. 2023 Dec 12;13(1):22049. doi: 10.1038/s41598-023-49049-6. Sci Rep. 2023. PMID: 38087002 Free PMC article.
References
-
- Baker D. M., Jordán-Dahlgren E., Maldonado M. A., Harvell C. D. (2010). Sea fan corals provide a stable isotope baseline for assessing sewage pollution in the Mexican Caribbean. Limnol. Oceanogr. 55 2139–2149. 10.4319/lo.2010.55.5.2139 - DOI
-
- Baker D. M., Rodriguez-Martinez R. E., Fogel M. L. (2013). Tourism’s nitrogen footprint on a Mesoamerican coral reef. Coral Reefs 32 691–699. 10.1007/s00338-013-1040-2 - DOI
Grants and funding
LinkOut - more resources
Full Text Sources

