Agent-Based Models Predict Emergent Behavior of Heterogeneous Cell Populations in Dynamic Microenvironments
- PMID: 32596213
- PMCID: PMC7301008
- DOI: 10.3389/fbioe.2020.00249
Agent-Based Models Predict Emergent Behavior of Heterogeneous Cell Populations in Dynamic Microenvironments
Abstract
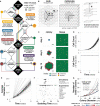
Computational models are most impactful when they explain and characterize biological phenomena that are non-intuitive, unexpected, or difficult to study experimentally. Countless equation-based models have been built for these purposes, but we have yet to realize the extent to which rules-based models offer an intuitive framework that encourages computational and experimental collaboration. We develop ARCADE, a multi-scale agent-based model to interrogate emergent behavior of heterogeneous cell agents within dynamic microenvironments and demonstrate how complexity of intracellular metabolism and signaling modules impacts emergent dynamics. We perform in silico case studies on context, competition, and heterogeneity to demonstrate the utility of our model for gaining computational and experimental insight. Notably, there exist (i) differences in emergent behavior between colony and tissue contexts, (ii) linear, non-linear, and multimodal consequences of parameter variation on competition in simulated co-cultures, and (iii) variable impact of cell and population heterogeneity on emergent outcomes. Our extensible framework is easily modified to explore numerous biological systems, from tumor microenvironments to microbiomes.
Keywords: agent-based model; cell population dynamics; computational modeling; emergent behavior; microenvironment.
Copyright © 2020 Yu and Bagheri.
Figures





References
-
- Alley M. C., Scudiero D. A., Monks A., Hursey M. L., Czerwinski M. J., Fine D. L., et al. . (1988). Feasibility of drug screening with panels of human tumor cell lines using a microculture tetrazolium assay. Cancer Res. 48, 589–601. - PubMed
LinkOut - more resources
Full Text Sources

