Sars-CoV-2 Envelope and Membrane Proteins: Structural Differences Linked to Virus Characteristics?
- PMID: 32596311
- PMCID: PMC7261327
- DOI: 10.1155/2020/4389089
Sars-CoV-2 Envelope and Membrane Proteins: Structural Differences Linked to Virus Characteristics?
Abstract
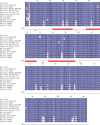
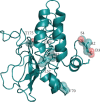
The Coronavirus Disease 2019 (COVID-19) is a new viral infection caused by the severe acute respiratory coronavirus 2 (SARS-CoV-2). Genomic analyses have revealed that SARS-CoV-2 is related to Pangolin and Bat coronaviruses. In this report, a structural comparison between the Sars-CoV-2 Envelope and Membrane proteins from different human isolates with homologous proteins from closely related viruses is described. The analyses here reported show the high structural similarity of Envelope and Membrane proteins to the counterparts from Pangolin and Bat coronavirus isolates. However, the comparisons have also highlighted structural differences specific of Sars-CoV-2 proteins which may be correlated to the cross-species transmission and/or to the properties of the virus. Structural modelling has been applied to map the variant sites onto the predicted three-dimensional structure of the Envelope and Membrane proteins.
Copyright © 2020 Martina Bianchi et al.
Conflict of interest statement
The authors declare that there is no conflict of interest regarding the publication of this paper.
Figures

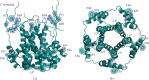
References
-
- Lai C.-C., Shih T.-P., Ko W.-C., Tang H.-J., Hsueh P.-R. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and coronavirus Disease-2019 (COVID-19): the epidemic and the challenges. International Journal of Antimicrobial Agents. 2020;55(3, article 105924) doi: 10.1016/j.ijantimicag.2020.105924. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

