Guide-free Cas9 from pathogenic Campylobacter jejuni bacteria causes severe damage to DNA
- PMID: 32596446
- PMCID: PMC7299616
- DOI: 10.1126/sciadv.aaz4849
Guide-free Cas9 from pathogenic Campylobacter jejuni bacteria causes severe damage to DNA
Abstract
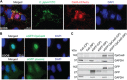
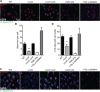
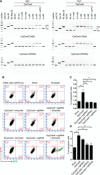
CRISPR-Cas9 systems are enriched in human pathogenic bacteria and have been linked to cytotoxicity by an unknown mechanism. Here, we show that upon infection of human cells, Campylobacter jejuni secretes its Cas9 (CjeCas9) nuclease into their cytoplasm. Next, a native nuclear localization signal enables CjeCas9 nuclear entry, where it catalyzes metal-dependent nonspecific DNA cleavage leading to cell death. Compared to CjeCas9, native Cas9 of Streptococcus pyogenes (SpyCas9) is more suitable for guide-dependent editing. However, in human cells, native SpyCas9 may still cause some DNA damage, most likely because of its ssDNA cleavage activity. This side effect can be completely prevented by saturation of SpyCas9 with an appropriate guide RNA, which is only partially effective for CjeCas9. We conclude that CjeCas9 plays an active role in attacking human cells rather than in viral defense. Moreover, these unique catalytic features may therefore make CjeCas9 less suitable for genome editing applications.
Copyright © 2020 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works. Distributed under a Creative Commons Attribution NonCommercial License 4.0 (CC BY-NC).
Figures

References
-
- Mohanraju P., Makarova K. S., Zetsche B., Zhang F., Koonin E. V., van der Oost J., Diverse evolutionary roots and mechanistic variations of the CRISPR-Cas systems. Science 353, aad5147 (2016). - PubMed
-
- Westra E. R., Buckling A., Fineran P. C., CRISPR-Cas systems: Beyond adaptive immunity. Nat. Rev. Microbiol. 12, 317–326 (2014). - PubMed
-
- Louwen R., Horst-Kreft D., Boer A. G., Graaf L., Knegt G., Hamersma M., Heikema A. P., Timms A. R., Jacobs B. C., Wagenaar J. A., Endtz H. P., Oost J., Wells J. M., Nieuwenhuis E. E. S., Vliet A. H. M., Willemsen P. T. J., Baarlen P., Belkum A., A novel link between Campylobacter jejuni bacteriophage defence, virulence and Guillain–Barré syndrome. Eur. J. Clin. Microbiol. Infect. Dis. 32, 207–226 (2013). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

