Evidence for host-dependent RNA editing in the transcriptome of SARS-CoV-2
- PMID: 32596474
- PMCID: PMC7299625
- DOI: 10.1126/sciadv.abb5813
Evidence for host-dependent RNA editing in the transcriptome of SARS-CoV-2
Abstract
The COVID-19 outbreak has become a global health risk, and understanding the response of the host to the SARS-CoV-2 virus will help to combat the disease. RNA editing by host deaminases is an innate restriction process to counter virus infection, but it is not yet known whether this process operates against coronaviruses. Here, we analyze RNA sequences from bronchoalveolar lavage fluids obtained from coronavirus-infected patients. We identify nucleotide changes that may be signatures of RNA editing: adenosine-to-inosine changes from ADAR deaminases and cytosine-to-uracil changes from APOBEC deaminases. Mutational analysis of genomes from different strains of Coronaviridae from human hosts reveals mutational patterns consistent with those observed in the transcriptomic data. However, the reduced ADAR signature in these data raises the possibility that ADARs might be more effective than APOBECs in restricting viral propagation. Our results thus suggest that both APOBECs and ADARs are involved in coronavirus genome editing, a process that may shape the fate of both virus and patient.
Copyright © 2020 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works. Distributed under a Creative Commons Attribution NonCommercial License 4.0 (CC BY-NC).
Figures
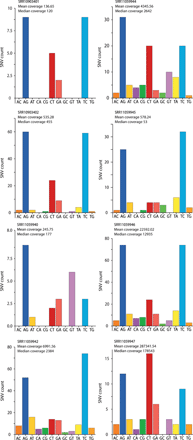
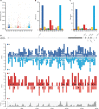
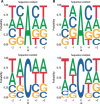
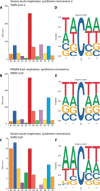
Comment in
-
Reconciling the debate on deamination on viral RNA.J Appl Genet. 2022 Sep;63(3):583-585. doi: 10.1007/s13353-022-00698-9. Epub 2022 May 4. J Appl Genet. 2022. PMID: 35507138 Free PMC article. No abstract available.
References
-
- Wu F., Zhao S., Yu B., Chen Y.-M., Wang W., Song Z.-G., Hu Y., Tao Z.-W., Tian J.-H., Pei Y.-Y., Yuan M.-L., Zhang Y.-L., Dai F.-H., Liu Y., Wang Q.-M., Zheng J.-J., Xu L., Holmes E. C., Zhang Y.-Z., A new coronavirus associated with human respiratory disease in China. Nature 579, 265–269 (2020). - PMC - PubMed
-
- Li Q., Guan X., Wu P., Wang X., Zhou L., Tong Y., Ren R., Leung K. S. M., Lau E. H. Y., Wong J. Y., Xing X., Xiang N., Wu Y., Li C., Chen Q., Li D., Liu T., Zhao J., Liu M., Tu W., Chen C., Jin L., Yang R., Wang Q., Zhou S., Wang R., Liu H., Luo Y., Liu Y., Shao G., Li H., Tao Z., Yang Y., Deng Z., Liu B., Ma Z., Zhang Y., Shi G., Lam T. T. Y., Wu J. T., Gao G. F., Cowling B. J., Yang B., Leung G. M., Feng Z., Early transmission dynamics in Wuhan, China, of novel coronavirus-infected pneumonia. N. Engl. J. Med. 382, 1199–1207 (2020). - PMC - PubMed
-
- Eisenberg E., Levanon E. Y., A-to-I RNA editing—Immune protector and transcriptome diversifier. Nat. Rev. Genet. 19, 473–490 (2018). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

