Application of serine integrases for secondary metabolite pathway assembly in Streptomyces
- PMID: 32596521
- PMCID: PMC7306541
- DOI: 10.1016/j.synbio.2020.05.006
Application of serine integrases for secondary metabolite pathway assembly in Streptomyces
Erratum in
-
Erratum regarding previously published articles.Synth Syst Biotechnol. 2020 Oct 14;5(4):330-331. doi: 10.1016/j.synbio.2020.10.001. eCollection 2020 Dec. Synth Syst Biotechnol. 2020. PMID: 33102827 Free PMC article.
Abstract
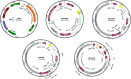
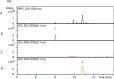
Serine integrases have been shown to be efficient tools for metabolic pathway assembly. To further improve the flexibility and efficiency of pathway engineering via serine integrases, we explored how multiple orthogonally active serine integrases can be applied for use in vitro for the heterologous expression of complex biosynthesis pathways in Streptomyces spp., the major producers of useful bioactive natural products. The results show that multiple orthogonal serine integrases efficiently assemble the genes from a complex biosynthesis pathway in a single in vitro recombination reaction, potentially permitting a versatile combinatorial assembly approach. Furthermore, the assembly strategy also permitted the incorporation of a well-characterised promoter upstream of each gene for expression in a heterologous host. The results demonstrate how site-specific recombination based on orthogonal serine integrases can be applied in Streptomyces spp.
Keywords: Erythromycin; Pathway assembly; Serine integrase; Streptomyces.
© 2020 KeAi Communications Co.(+) Ltd.
Figures



References
-
- Sosio M. Artificial chromosomes for antibiotic-producing actinomycetes. Nat Biotechnol. 2000;18(3):343–345. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

