The Human Gene Mutation Database (HGMD®): optimizing its use in a clinical diagnostic or research setting
- PMID: 32596782
- PMCID: PMC7497289
- DOI: 10.1007/s00439-020-02199-3
The Human Gene Mutation Database (HGMD®): optimizing its use in a clinical diagnostic or research setting
Abstract
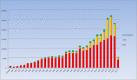
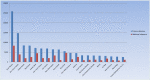
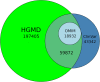
The Human Gene Mutation Database (HGMD®) constitutes a comprehensive collection of published germline mutations in nuclear genes that are thought to underlie, or are closely associated with human inherited disease. At the time of writing (June 2020), the database contains in excess of 289,000 different gene lesions identified in over 11,100 genes manually curated from 72,987 articles published in over 3100 peer-reviewed journals. There are primarily two main groups of users who utilise HGMD on a regular basis; research scientists and clinical diagnosticians. This review aims to highlight how to make the most out of HGMD data in each setting.
Conflict of interest statement
The authors wish to declare an interest insofar as HGMD is financially supported by Qiagen Inc. through a License agreement with Cardiff University.
Figures


References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

