A CLN6-CLN8 complex recruits lysosomal enzymes at the ER for Golgi transfer
- PMID: 32597833
- PMCID: PMC7410054
- DOI: 10.1172/JCI130955
A CLN6-CLN8 complex recruits lysosomal enzymes at the ER for Golgi transfer
Abstract
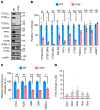
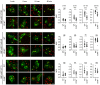
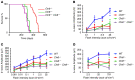
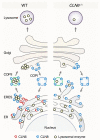
Lysosomal enzymes are synthesized in the endoplasmic reticulum (ER) and transferred to the Golgi complex by interaction with the Batten disease protein CLN8 (ceroid lipofuscinosis, neuronal, 8). Here we investigated the relationship of this pathway with CLN6, an ER-associated protein of unknown function that is defective in a different Batten disease subtype. Experiments focused on protein interaction and trafficking identified CLN6 as an obligate component of a CLN6-CLN8 complex (herein referred to as EGRESS: ER-to-Golgi relaying of enzymes of the lysosomal system), which recruits lysosomal enzymes at the ER to promote their Golgi transfer. Mutagenesis experiments showed that the second luminal loop of CLN6 is required for the interaction of CLN6 with the enzymes but dispensable for interaction with CLN8. In vitro and in vivo studies showed that CLN6 deficiency results in inefficient ER export of lysosomal enzymes and diminished levels of the enzymes at the lysosome. Mice lacking both CLN6 and CLN8 did not display aggravated pathology compared with the single deficiencies, indicating that the EGRESS complex works as a functional unit. These results identify CLN6 and the EGRESS complex as key players in lysosome biogenesis and shed light on the molecular etiology of Batten disease caused by defects in CLN6.
Keywords: Cell Biology; Genetic diseases; Lysosomes; Molecular pathology.
Conflict of interest statement
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

