Epithelial-derived gasdermin D mediates nonlytic IL-1β release during experimental colitis
- PMID: 32597834
- PMCID: PMC7410065
- DOI: 10.1172/JCI138103
Epithelial-derived gasdermin D mediates nonlytic IL-1β release during experimental colitis
Abstract
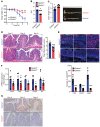
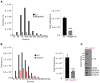
Gasdermin D (GSDMD) induces pyroptosis via the pore-forming activity of its N-terminal domain, cleaved by activated caspases associated with the release of IL-1β. Here, we report a nonpyroptotic role of full-length GSDMD in guiding the release of IL-1β-containing small extracellular vesicles (sEVs) from intestinal epithelial cells (IECs). In response to caspase-8 inflammasome activation, GSDMD, chaperoned by Cdc37/Hsp90, recruits the E3 ligase, NEDD4, to catalyze polyubiquitination of pro-IL-1β, serving as a signal for cargo loading into secretory vesicles. GSDMD and IL-1β colocalize with the exosome markers CD63 and ALIX intracellularly, and GSDMD and NEDD4 are required for release of CD63+ sEVs containing IL-1β, GSDMD, NEDD4, and caspase-8. Importantly, increased expression of epithelial-derived GSDMD is observed both in patients with inflammatory bowel disease (IBD) and those with experimental colitis. While GSDMD-dependent release of IL-1β-containing sEVs is detected in cultured colonic explants from colitic mice, GSDMD deficiency substantially attenuates disease severity, implicating GSDMD-mediated release of IL-1β sEVs in the pathogenesis of intestinal inflammation, such as that observed in IBD.
Keywords: Cytokines; Gastroenterology; Inflammation; Inflammatory bowel disease; Innate immunity.
Conflict of interest statement
Figures








References
Publication types
MeSH terms
Substances
Grants and funding
- S10 RR031537/RR/NCRR NIH HHS/United States
- MC_PC_MR/S025952/1/MRC_/Medical Research Council/United Kingdom
- P01 HL103453/HL/NHLBI NIH HHS/United States
- R01 DK042191/DK/NIDDK NIH HHS/United States
- MR/S036377/1/MRC_/Medical Research Council/United Kingdom
- MC_UU_00008/7/MRC_/Medical Research Council/United Kingdom
- R01 DK055812/DK/NIDDK NIH HHS/United States
- MC_UU_12010/7/MRC_/Medical Research Council/United Kingdom
- T32 GM007250/GM/NIGMS NIH HHS/United States
- P01 DK091222/DK/NIDDK NIH HHS/United States
- S10 OD023436/OD/NIH HHS/United States
- P30 DK097948/DK/NIDDK NIH HHS/United States
- R01 NS104164/NS/NINDS NIH HHS/United States
- P01 HL029582/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

