Interferon-stimulated gene 15 accelerates replication fork progression inducing chromosomal breakage
- PMID: 32597933
- PMCID: PMC7401800
- DOI: 10.1083/jcb.202002175
Interferon-stimulated gene 15 accelerates replication fork progression inducing chromosomal breakage
Abstract
DNA replication is highly regulated by the ubiquitin system, which plays key roles upon stress. The ubiquitin-like modifier ISG15 (interferon-stimulated gene 15) is induced by interferons, bacterial and viral infection, and DNA damage, but it is also constitutively expressed in many types of cancer, although its role in tumorigenesis is still largely elusive. Here, we show that ISG15 localizes at the replication forks, in complex with PCNA and the nascent DNA, where it regulates DNA synthesis. Indeed, high levels of ISG15, intrinsic or induced by interferon-β, accelerate DNA replication fork progression, resulting in extensive DNA damage and chromosomal aberrations. This effect is largely independent of ISG15 conjugation and relies on ISG15 functional interaction with the DNA helicase RECQ1, which promotes restart of stalled replication forks. Additionally, elevated ISG15 levels sensitize cells to cancer chemotherapeutic treatments. We propose that ISG15 up-regulation exposes cells to replication stress, impacting genome stability and response to genotoxic drugs.
© 2020 Raso et al.
Figures
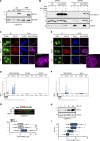




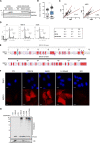

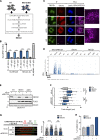
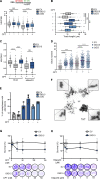
Comment in
-
ISG15 fast-tracks DNA replication.J Cell Biol. 2020 Aug 3;219(8):e202007028. doi: 10.1083/jcb.202007028. J Cell Biol. 2020. PMID: 32687143 Free PMC article.
References
-
- Bektas N., Noetzel E., Veeck J., Press M.F., Kristiansen G., Naami A., Hartmann A., Dimmler A., Beckmann M.W., Knüchel R., et al. . 2008. The ubiquitin-like molecule interferon-stimulated gene 15 (ISG15) is a potential prognostic marker in human breast cancer. Breast Cancer Res. 10:R58 10.1186/bcr2117 - DOI - PMC - PubMed
-
- Berti M., Ray Chaudhuri A., Thangavel S., Gomathinayagam S., Kenig S., Vujanovic M., Odreman F., Glatter T., Graziano S., Mendoza-Maldonado R., et al. . 2013. Human RECQ1 promotes restart of replication forks reversed by DNA topoisomerase I inhibition. Nat. Struct. Mol. Biol. 20:347–354. 10.1038/nsmb.2501 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

