MicroRNA-5572 Is a Novel MicroRNA-Regulating SLC30A3 in Sporadic Amyotrophic Lateral Sclerosis
- PMID: 32599739
- PMCID: PMC7350020
- DOI: 10.3390/ijms21124482
MicroRNA-5572 Is a Novel MicroRNA-Regulating SLC30A3 in Sporadic Amyotrophic Lateral Sclerosis
Abstract
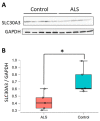
Amyotrophic lateral sclerosis (ALS) is a progressive degenerative disease caused by the loss of motor neurons. Although the pathogenesis of sporadic ALS (sALS) remains unclear, it has recently been suggested that disorders of microRNA (miRNA) may be involved in neurodegenerative conditions. The purpose of this study was to investigate miRNA levels in sALS and the target genes of miRNA. Microarray and real-time RT-PCR analyses revealed significantly-decreased levels of miR-139-5p and significantly increased levels of miR-5572 in the spinal cords of sALS patients compared with those in controls. We then focused on miR-5572, which has not been reported in ALS, and determined its target gene. By using TargetScan, we predicted SLC30A3 as the candidate target gene of miR-5572. In a previous study, we found decreased SLC30A3 levels in the spinal cords of sALS patients. We revealed that SLC30A3 was regulated by miR-5572. Taken together, these results demonstrate that the level of novel miRNA miR-5572 is increased in sALS and that SLC30A3 is one of the target genes regulated by miR-5572.
Keywords: amyotrophic lateral sclerosis; microRNA; microarray; spinal cord.
Conflict of interest statement
The authors declare that there is no conflict of interests.
Figures



References
-
- Kaneko M., Noguchi T., Ikegami S., Sakurai T., Kakita A., Toyoshima Y., Kambe T., Yamada M., Inden M., Hara H., et al. Zinc transporters ZnT3 and ZnT6 are downregulated in the spinal cords of patients with sporadic amyotrophic lateral sclerosis. J. Neurosci. Res. 2015;93:370–379. doi: 10.1002/jnr.23491. - DOI - PubMed
MeSH terms
Substances
Grants and funding
- 15K21278/Japan Society for the Promotion of Science
- 17K18001/Japan Society for the Promotion of Science
- JP19H05767A02/Japan Society for the Promotion of Science
- 2019-23/the Collaborative Research Project of Brain Research Institute, Niigata University
- 28-16/Grant-in-Aid of The Nakabayashi Trust For ALS Research
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

