Genetic Characterization of Native Donkey (Equus asinus) Populations of Turkey Using Microsatellite Markers
- PMID: 32599857
- PMCID: PMC7341297
- DOI: 10.3390/ani10061093
Genetic Characterization of Native Donkey (Equus asinus) Populations of Turkey Using Microsatellite Markers
Abstract
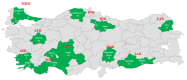
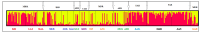
This study presents the first insights to the genetic diversity and structure of the Turkish donkey populations. The primary objectives were to detect the main structural features of Turkish donkeys by microsatellite markers. A panel of 17 microsatellite markers was applied for genotyping 314 donkeys from 16 locations of Turkey. One hundred and forty-two alleles were identified and the number of alleles per locus ranged from 4 to 12. The highest number of alleles was observed in AHT05 (12) and the lowest in ASB02 and HTG06 (4), while ASB17 was monomorphic. The mean HO in the Turkish donkey was estimated to be 0.677, while mean HE was 0.675. The polymorphic information content (PIC) was calculated for each locus and ranged from 0.36 (locus ASB02) to 0.98 (locus AHT05), which has the highest number of alleles per locus in the present study. The average PIC in our populations was 0.696. The average coefficient of gene differentiation (GST) over the 17 loci was 0.020 ± 0.037 (p < 0.01). The GST values for single loci ranged from -0.004 for LEX54 to 0.162 for COR082. Nei's gene diversity index (Ht) for loci ranged from 0.445 (ASB02) to 0.890 (AHT05), with an average of 0.696. A Bayesian clustering method, the Structure software, was used for clustering algorithms of multi-locus genotypes to identify the population structure and the pattern of admixture within the populations. When the number of ancestral populations varied from K = 1 to 20, the largest change in the log of the likelihood function (K) was when K = 2. The results for K = 2 indicate a clear separation between Clade I (KIR, CAT, KAR, MAR, SAN) and Clade II (MAL, MER, TOK, KAS, KUT, KON, ISP, ANT, MUG, AYD and KAH) populations.
Keywords: Equus asinus; Turkey; donkey; genetic diversity; microsatellites; molecular markers.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Mittermeier R.A., Gil P.R., Hoffman M., Pilgrim J., Brooks T., Mittermeier C.G. Hotspots Revisited: Earth’s Biologically Richest and Most Endangered Terrestrial Ecoregions. Conserv. Int. 2005;392
-
- Şekercioğlu C.H., Anderson S., Akcay E., Bilgin R., Can O.E., Semiz G., Tavşanoğlu C., Yokeş M.B., Soyumert A., Ipekdal K., et al. Turkey’s globally important biodiversity in crisis. Boil. Conserv. 2011;144:2752–2769. doi: 10.1016/j.biocon.2011.06.025. - DOI
-
- Kugler W., Grünenfelder H.-P., Broxham E. Donkey Breeds in Europe. Inventory, Description, Need for Action: Report. Nat. Sustain. 2018:679–685.
-
- Yilmaz O., Ertuğrul M. Domestication of Donkey (Equus Asinus). Iğdır Üniversitesi Fen Bilim. Enstitüsü Derg. 2011;1:111–115.
-
- Yılmaz O., Wilson T.R. The Domestic Livestock Resources of Turkey: Notes on Donkeys. J. Anim. Plant. Sci. 2013;23:651–656.
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

