S1 Domain RNA-Binding Protein CvfD Is a New Posttranscriptional Regulator That Mediates Cold Sensitivity, Phosphate Transport, and Virulence in Streptococcus pneumoniae D39
- PMID: 32601068
- PMCID: PMC7925082
- DOI: 10.1128/JB.00245-20
S1 Domain RNA-Binding Protein CvfD Is a New Posttranscriptional Regulator That Mediates Cold Sensitivity, Phosphate Transport, and Virulence in Streptococcus pneumoniae D39
Abstract
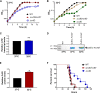
Posttranscriptional gene regulation often involves RNA-binding proteins that modulate mRNA translation and/or stability either directly through protein-RNA interactions or indirectly by facilitating the annealing of small regulatory RNAs (sRNAs). The human pathogen Streptococcus pneumoniae D39 (pneumococcus) does not encode homologs to RNA-binding proteins known to be involved in promoting sRNA stability and function, such as Hfq or ProQ, even though it contains genes for at least 112 sRNAs. However, the pneumococcal genome contains genes for other RNA-binding proteins, including at least six S1 domain proteins: ribosomal protein S1 (rpsA), polynucleotide phosphorylase (pnpA), RNase R (rnr), and three proteins with unknown functions. Here, we characterize the function of one of these conserved, yet uncharacterized, S1 domain proteins, SPD_1366, which we have renamed CvfD (
Keywords: RNA-binding protein; cold sensitivity; phosphate uptake; pleiotropic regulation; pneumococcus; posttranscriptional regulation; virulence attenuation.
Copyright © 2020 American Society for Microbiology.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

