A Structure-Function-Inhibition Analysis of the Pseudomonas aeruginosa Type III Secretion Needle Protein PscF
- PMID: 32601072
- PMCID: PMC7925083
- DOI: 10.1128/JB.00055-20
A Structure-Function-Inhibition Analysis of the Pseudomonas aeruginosa Type III Secretion Needle Protein PscF
Abstract
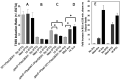
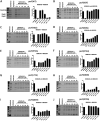
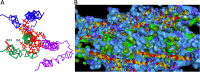
The Pseudomonas aeruginosa type III secretion system (T3SS) needle comprised of multiple PscF subunits is essential for the translocation of effector toxins into human cells, facilitating the establishment and dissemination of infection. Mutations in the pscF gene provide resistance to the phenoxyacetamide (PhA) series of T3SS inhibitory chemical probes. To better understand PscF functions and interactions with PhA, alleles of pscF with 71 single mutations altering 49 of the 85 residues of the encoded protein were evaluated for their effects on T3SS phenotypes. Of these, 37% eliminated and 63% maintained secretion, with representatives of both evenly distributed across the entire protein. Mutations in 14 codons conferred a degree of PhA resistance without eliminating secretion, and all but one were in the alpha-helical C-terminal 25% of PscF. PhA-resistant mutants exhibited no cross-resistance to two T3SS inhibitors with different chemical scaffolds. Two mutations caused constitutive T3SS secretion. The pscF allele at its native locus, whether wild type (WT), constitutive, or PhA resistant, was dominant over other pscF alleles expressed from nonnative loci and promoters, but mixed phenotypes were observed in chromosomal ΔpscF strains with both WT and mutant alleles at nonnative loci. Some PhA-resistant mutants exhibited reduced translocation efficiency that was improved in a PhA dose-dependent manner, suggesting that PhA can bind to those resistant needles. In summary, these results are consistent with a direct interaction between PhA inhibitors and the T3SS needle, suggest a mechanism of blocking conformational changes, and demonstrate that PscF affects T3SS regulation, as well as carrying out secretion and translocation.IMPORTANCEP. aeruginosa effector toxin translocation into host innate immune cells is critical for the establishment and dissemination of P. aeruginosa infections. The medical need for new anti-P. aeruginosa agents is evident by the fact that P. aeruginosa ventilator-associated pneumonia is associated with a high mortality rate (40 to 69%) and recurs in >30% of patients, even with standard-of-care antibiotic therapy. The results described here confirm roles for the PscF needle in T3SS secretion and translocation and suggest that it affects regulation, possibly by interaction with T3SS regulatory proteins. The results also support a model of direct interaction of the needle with PhA and suggest that, with further development, members of the PhA series may prove useful as drugs for P. aeruginosa infection.
Keywords: P. aeruginosa; phenoxyacetamides; translocation; type III needle protein; type III secretion.
Copyright © 2020 American Society for Microbiology.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

