The Arabidopsis epigenetic regulator ICU11 as an accessory protein of Polycomb Repressive Complex 2
- PMID: 32601198
- PMCID: PMC7368280
- DOI: 10.1073/pnas.1920621117
The Arabidopsis epigenetic regulator ICU11 as an accessory protein of Polycomb Repressive Complex 2
Abstract
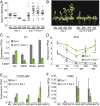
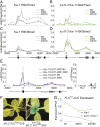
Molecular mechanisms enabling the switching and maintenance of epigenetic states are not fully understood. Distinct histone modifications are often associated with ON/OFF epigenetic states, but how these states are stably maintained through DNA replication, yet in certain situations switch from one to another remains unclear. Here, we address this problem through identification of Arabidopsis INCURVATA11 (ICU11) as a Polycomb Repressive Complex 2 accessory protein. ICU11 robustly immunoprecipitated in vivo with PRC2 core components and the accessory proteins, EMBRYONIC FLOWER 1 (EMF1), LIKE HETEROCHROMATIN PROTEIN1 (LHP1), and TELOMERE_REPEAT_BINDING FACTORS (TRBs). ICU11 encodes a 2-oxoglutarate-dependent dioxygenase, an activity associated with histone demethylation in other organisms, and mutant plants show defects in multiple aspects of the Arabidopsis epigenome. To investigate its primary molecular function we identified the Arabidopsis FLOWERING LOCUS C (FLC) as a direct target and found icu11 disrupted the cold-induced, Polycomb-mediated silencing underlying vernalization. icu11 prevented reduction in H3K36me3 levels normally seen during the early cold phase, supporting a role for ICU11 in H3K36me3 demethylation. This was coincident with an attenuation of H3K27me3 at the internal nucleation site in FLC, and reduction in H3K27me3 levels across the body of the gene after plants were returned to the warm. Thus, ICU11 is required for the cold-induced epigenetic switching between the mutually exclusive chromatin states at FLC, from the active H3K36me3 state to the silenced H3K27me3 state. These data support the importance of physical coupling of histone modification activities to promote epigenetic switching between opposing chromatin states.
Keywords: ICU11; Polycomb; chromatin; epigenetic.
Copyright © 2020 the Author(s). Published by PNAS.
Conflict of interest statement
The authors declare no competing interest.
Figures


References
-
- Grossniklaus U., Vielle-Calzada J. P., Hoeppner M. A., Gagliano W. B., Maternal control of embryogenesis by MEDEA, a polycomb group gene in Arabidopsis. Science 280, 446–450 (1998). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- 203149/WT_/Wellcome Trust/United Kingdom
- BBS/E/J/00000581/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- 108504/WT_/Wellcome Trust/United Kingdom
- BB/J004588/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/S009620/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

