Mechanisms underlying homeostatic plasticity in the Drosophila mushroom body in vivo
- PMID: 32601210
- PMCID: PMC7368247
- DOI: 10.1073/pnas.1921294117
Mechanisms underlying homeostatic plasticity in the Drosophila mushroom body in vivo
Abstract
Neural network function requires an appropriate balance of excitation and inhibition to be maintained by homeostatic plasticity. However, little is known about homeostatic mechanisms in the intact central brain in vivo. Here, we study homeostatic plasticity in the Drosophila mushroom body, where Kenyon cells receive feedforward excitation from olfactory projection neurons and feedback inhibition from the anterior paired lateral neuron (APL). We show that prolonged (4-d) artificial activation of the inhibitory APL causes increased Kenyon cell odor responses after the artificial inhibition is removed, suggesting that the mushroom body compensates for excess inhibition. In contrast, there is little compensation for lack of inhibition (blockade of APL). The compensation occurs through a combination of increased excitation of Kenyon cells and decreased activation of APL, with differing relative contributions for different Kenyon cell subtypes. Our findings establish the fly mushroom body as a model for homeostatic plasticity in vivo.
Keywords: Drosophila; homeostatic plasticity; mushroom body; olfaction.
Conflict of interest statement
The authors declare no competing interest.
Figures

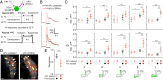
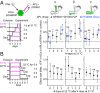
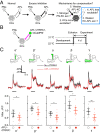

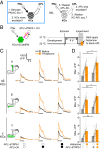
References
-
- Olshausen B. A., Field D. J., Sparse coding of sensory inputs. Curr. Opin. Neurobiol. 14, 481–487 (2004). - PubMed
-
- Litwin-Kumar A., Doiron B., Formation and maintenance of neuronal assemblies through synaptic plasticity. Nat. Commun. 5, 5319 (2014). - PubMed
-
- Keck T., Hübener M., Bonhoeffer T., Interactions between synaptic homeostatic mechanisms: An attempt to reconcile BCM theory, synaptic scaling, and changing excitation/inhibition balance. Curr. Opin. Neurobiol. 43, 87–93 (2017). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

