Yaravirus: A novel 80-nm virus infecting Acanthamoeba castellanii
- PMID: 32601223
- PMCID: PMC7368276
- DOI: 10.1073/pnas.2001637117
Yaravirus: A novel 80-nm virus infecting Acanthamoeba castellanii
Abstract
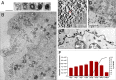
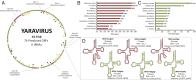
Here we report the discovery of Yaravirus, a lineage of amoebal virus with a puzzling origin and evolution. Yaravirus presents 80-nm-sized particles and a 44,924-bp dsDNA genome encoding for 74 predicted proteins. Yaravirus genome annotation showed that none of its genes matched with sequences of known organisms at the nucleotide level; at the amino acid level, six predicted proteins had distant matches in the nr database. Complimentary prediction of three-dimensional structures indicated possible function of 17 proteins in total. Furthermore, we were not able to retrieve viral genomes closely related to Yaravirus in 8,535 publicly available metagenomes spanning diverse habitats around the globe. The Yaravirus genome also contained six types of tRNAs that did not match commonly used codons. Proteomics revealed that Yaravirus particles contain 26 viral proteins, one of which potentially representing a divergent major capsid protein (MCP) with a predicted double jelly-roll domain. Structure-guided phylogeny of MCP suggests that Yaravirus groups together with the MCPs of Pleurochrysis endemic viruses. Yaravirus expands our knowledge of the diversity of DNA viruses. The phylogenetic distance between Yaravirus and all other viruses highlights our still preliminary assessment of the genomic diversity of eukaryotic viruses, reinforcing the need for the isolation of new viruses of protists.
Keywords: NCLDV; ORFan; Yaravirus; capsid; metagenomics.
Conflict of interest statement
The authors declare no competing interest.
Figures



References
-
- Colson P., Ominami Y., Hisada A., La Scola B., Raoult D., Giant mimiviruses escape many canonical criteria of the virus definition. Clin. Microbiol. Infect. 25, 147–154 (2019). - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Miscellaneous

