Pre-existing immunity to influenza virus hemagglutinin stalk might drive selection for antibody-escape mutant viruses in a human challenge model
- PMID: 32601336
- PMCID: PMC7450362
- DOI: 10.1038/s41591-020-0937-x
Pre-existing immunity to influenza virus hemagglutinin stalk might drive selection for antibody-escape mutant viruses in a human challenge model
Abstract
The conserved region of influenza hemagglutinin (HA) stalk (or stem) has gained attention as a potent target for universal influenza vaccines1-5. Although the HA stalk region is relatively well conserved, the evolutionarily dynamic nature of influenza viruses6 raises concerns about the possible emergence of viruses carrying stalk escape mutation(s) under sufficient immune pressure. Here we show that immune pressure on the HA stalk can lead to expansion of escape mutant viruses in study participants challenged with a 2009 H1N1 pandemic influenza virus inoculum containing an A388V polymorphism in the HA stalk (45% wild type and 55% mutant). High level of stalk antibody titers was associated with the selection of the mutant virus both in humans and in vitro. Although the mutant virus showed slightly decreased replication in mice, it was not observed in cell culture, ferrets or human challenge participants. The A388V mutation conferred resistance to some of the potent HA stalk broadly neutralizing monoclonal antibodies (bNAbs). Co-culture of wild-type and mutant viruses in the presence of either a bNAb or human serum resulted in rapid expansion of the mutant. These data shed light on a potential obstacle for the success of HA-stalk-targeting universal influenza vaccines-viral escape from vaccine-induced stalk immunity.
Conflict of interest statement
Competing interests
All authors declare no competing financial or non-financial interests.
Figures

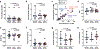
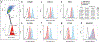







References
-
- Impagliazzo A et al. A stable trimeric influenza hemagglutinin stem as a broadly protective immunogen. Science 349, 1301–1306 (2015). - PubMed
-
- Yassine HM et al. Hemagglutinin-stem nanoparticles generate heterosubtypic influenza protection. Nat. Med 21, 1065–1070 (2015). - PubMed
-
- Andrews SF, Graham BS, Mascola JR & McDermott AB Is it possible to develop a “universal” influenza virus vaccine? Immunogenetic considerations underlying B-cell biology in the development of a pan-subtype influenza A vaccine targeting the hemagglutinin stem. Cold Spring Harb. Perspect. Biol 10, a029413 (2018). - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

