Multiclass cancer classification in fresh frozen and formalin-fixed paraffin-embedded tissue by DigiWest multiplex protein analysis
- PMID: 32601356
- PMCID: PMC7498367
- DOI: 10.1038/s41374-020-0455-y
Multiclass cancer classification in fresh frozen and formalin-fixed paraffin-embedded tissue by DigiWest multiplex protein analysis
Abstract
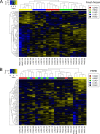
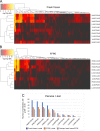
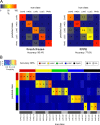
Histomorphology and immunohistochemistry are the most common ways of cancer classification in routine cancer diagnostics, but often reach their limits in determining the organ origin in metastasis. These cancers of unknown primary, which are mostly adenocarcinomas or squamous cell carcinomas, therefore require more sophisticated methodologies of classification. Here, we report a multiplex protein profiling-based approach for the classification of fresh frozen and formalin-fixed paraffin-embedded (FFPE) cancer tissue samples using the digital western blot technique DigiWest. A DigiWest-compatible FFPE extraction protocol was developed, and a total of 634 antibodies were tested in an initial set of 16 FFPE samples covering tumors from different origins. Of the 303 detected antibodies, 102 yielded significant correlation of signals in 25 pairs of fresh frozen and FFPE primary tumor samples, including head and neck squamous cell carcinomas (HNSC), lung squamous cell carcinomas (LUSC), lung adenocarcinomas (LUAD), colorectal adenocarcinomas (COAD), and pancreatic adenocarcinomas (PAAD). For this signature of 102 analytes (covering 88 total proteins and 14 phosphoproteins), a support vector machine (SVM) algorithm was developed. This allowed for the classification of the tissue of origin for all five tumor types studied here with high overall accuracies in both fresh frozen (90.4%) and FFPE (77.6%) samples. In addition, the SVM classifier reached an overall accuracy of 88% in an independent validation cohort of 25 FFPE tumor samples. Our results indicate that DigiWest-based protein profiling represents a valuable method for cancer classification, yielding conclusive and decisive data not only from fresh frozen specimens but also FFPE samples, thus making this approach attractive for routine clinical applications.
Conflict of interest statement
GE, JS, AA, and CS are employees of NMI TT Pharmaservices, a company offering DigiWest service studies. All other authors declare no conflicts of interest.
Figures

References
-
- Pavlidis N, Pentheroudakis G. Cancer of unknown primary site. Lancet. 2012;379:1428–35. - PubMed
-
- Economopoulou P, Mountzios G, Pavlidis N, Pentheroudakis G. Cancer of unknown primary origin in the genomic era: elucidating the dark box of cancer. Cancer Treat Rev. 2015;41:598–604. - PubMed
-
- Pentheroudakis G, Golfinopoulos V, Pavlidis N. Switching benchmarks in cancer of unknown primary: from autopsy to microarray. Eur J Cancer. 2007;43:2026–36. - PubMed
-
- Pereira TC, Share SM, Magalhães AV, Silverman JF. Can we tell the site of origin of metastatic squamous cell carcinoma? An immunohistochemical tissue microarray study of 194 cases. Appl Immunohistochem Mol Morphol. 2011;19:10–14. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

