Ancient familial Mediterranean fever mutations in human pyrin and resistance to Yersinia pestis
- PMID: 32601469
- PMCID: PMC7381377
- DOI: 10.1038/s41590-020-0705-6
Ancient familial Mediterranean fever mutations in human pyrin and resistance to Yersinia pestis
Abstract
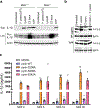
Familial Mediterranean fever (FMF) is an autoinflammatory disease caused by homozygous or compound heterozygous gain-of-function mutations in MEFV, which encodes pyrin, an inflammasome protein. Heterozygous carrier frequencies for multiple MEFV mutations are high in several Mediterranean populations, suggesting that they confer selective advantage. Among 2,313 Turkish people, we found extended haplotype homozygosity flanking FMF-associated mutations, indicating evolutionarily recent positive selection of FMF-associated mutations. Two pathogenic pyrin variants independently arose >1,800 years ago. Mutant pyrin interacts less avidly with Yersinia pestis virulence factor YopM than with wild-type human pyrin, thereby attenuating YopM-induced interleukin (IL)-1β suppression. Relative to healthy controls, leukocytes from patients with FMF harboring homozygous or compound heterozygous mutations and from asymptomatic heterozygous carriers released heightened IL-1β specifically in response to Y. pestis. Y. pestis-infected MefvM680I/M680I FMF knock-in mice exhibited IL-1-dependent increased survival relative to wild-type knock-in mice. Thus, FMF mutations that were positively selected in Mediterranean populations confer heightened resistance to Y. pestis.
Conflict of interest statement
COMPETING INTERESTS
The authors declare no competing interests
Figures




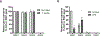
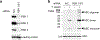






Comment in
-
Plague as a cause for familial Mediterranean fever.Nat Immunol. 2020 Aug;21(8):833-834. doi: 10.1038/s41590-020-0724-3. Nat Immunol. 2020. PMID: 32601468 No abstract available.
-
Pyrin variants can burn out the plague.Nat Rev Immunol. 2020 Aug;20(8):462-463. doi: 10.1038/s41577-020-0395-1. Nat Rev Immunol. 2020. PMID: 32636477 No abstract available.
References
-
- International_FMF_Consortium. Ancient missense mutations in a new member of the RoRet gene family are likely to cause familial Mediterranean fever. Cell 90, 797–807 (1997). - PubMed
-
- French_FMF_Consortium. A candidate gene for familial Mediterranean fever. Nat. Genet 17, 25–31 (1997). - PubMed
-
- Sohar E, Gafni J, Pras M & Heller H Familial Mediterranean fever. A survey of 470 cases and review of the literature. Am. J. Med 43, 227–253 (1967). - PubMed
-
- Touitou I The spectrum of Familial Mediterranean Fever (FMF) mutations. Eur. J. of Hum. Genet 9, 473–483 (2001). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

